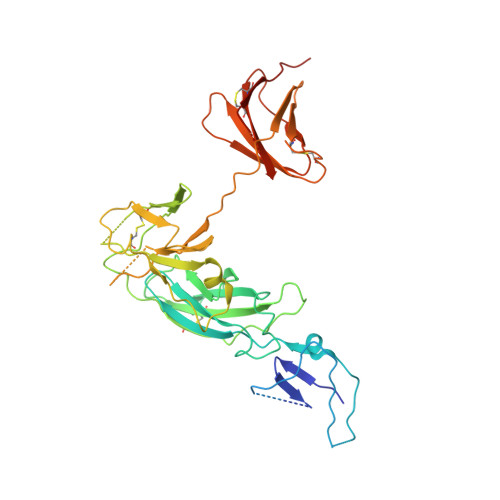
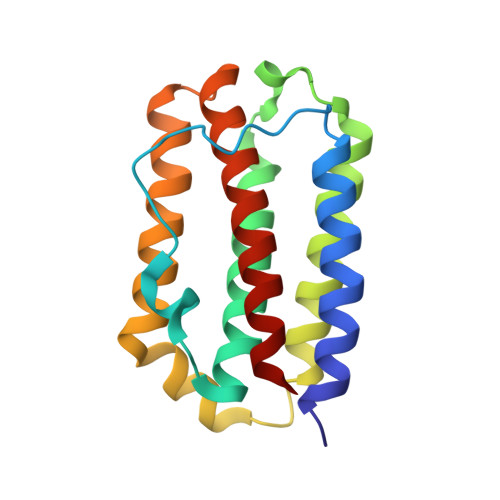
Structural basis of a unique interferon beta signaling axis mediated via the IFNAR1 receptor
de Weerd, N.A., Vivian, J.P., Nguyen, T.K., Mangan, N.E., Gould, J.A., Braniff, S.J., Zaker-Tabrizi, L., Fung, K.Y., Forster, S.C., Beddoe, T., Reid, H.H., Rossjohn, J., Hertzog, P.J.To be published.