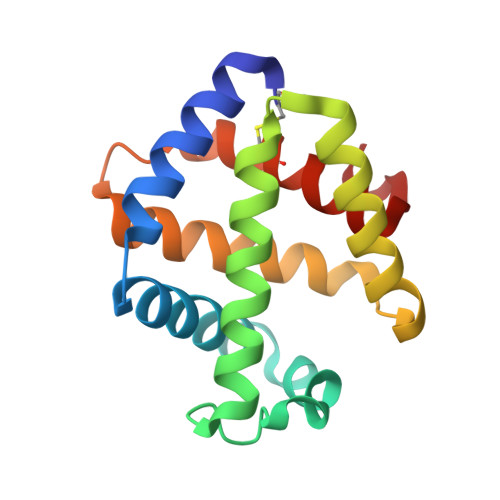
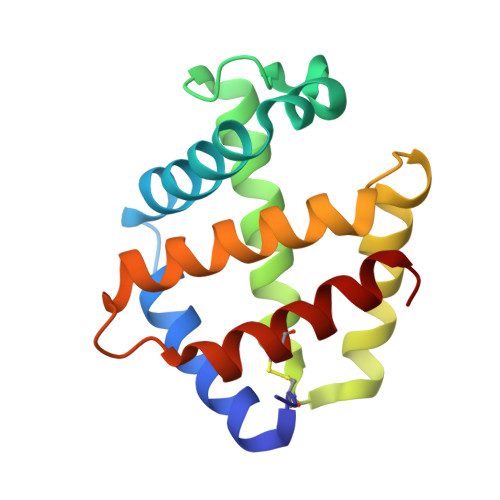
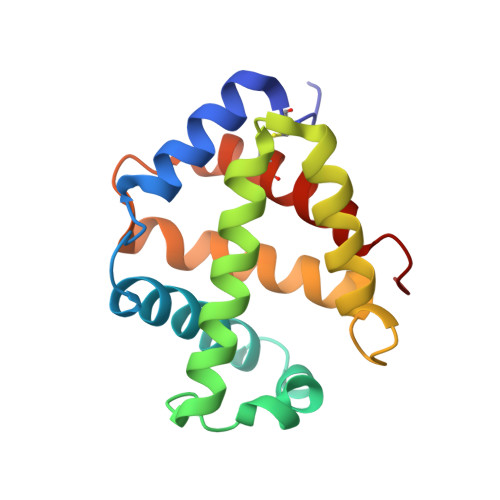
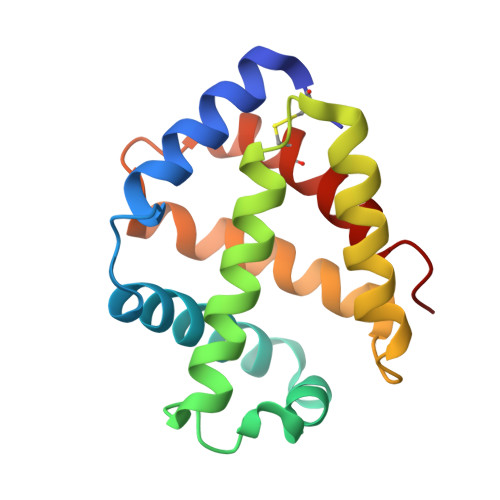
The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Numoto, N., Nakagawa, T., Ohara, R., Hasegawa, T., Kita, A., Yoshida, T., Maruyama, T., Imai, K., Fukumori, Y., Miki, K.(2014) Acta Crystallogr D Biol Crystallogr 70: 1823-1831
- PubMed: 25004960
- DOI: https://doi.org/10.1107/S1399004714008475
- Primary Citation of Related Structures:
3WCT, 3WCU, 3WCV, 3WCW - PubMed Abstract:
The quaternary structures of invertebrate haemoglobins (Hbs) are quite different from those of vertebrate Hbs. The extracellular giant Hbs of molecular masses of about 400 and 3600 kDa are composed of a dome-shaped dodecameric subassembly which consists of four individual globin subunits. Several crystal structures of 400 kDa Hbs from annelids have been reported, including structures in oxygenated and partially unliganded states, but the structure of the fully deoxygenated state has not been reported. In the present study, crystal structures of V2Hb from the tube worm Lamellibrachia satsuma have been determined in both the fully oxygenated and deoxygenated states. A glycosylation site and novel metal-binding sites for divalent cations were clearly observed with no intersubunit interactions in V2Hb. A comparison of the oxygenated and the deoxygenated forms of V2Hb reveals that the ternary- and quaternary-structural changes occur in a manner that maintains the molecular D3 symmetry. These structures suggest that the mechanisms of quaternary-structural changes between the oxy and deoxy states for the giant Hbs are identical across species.
- Department of Life Science, Graduate School of Natural Science and Technology, Kanazawa University, Kanazawa, Ishikawa 920-1192, Japan.
Organizational Affiliation: