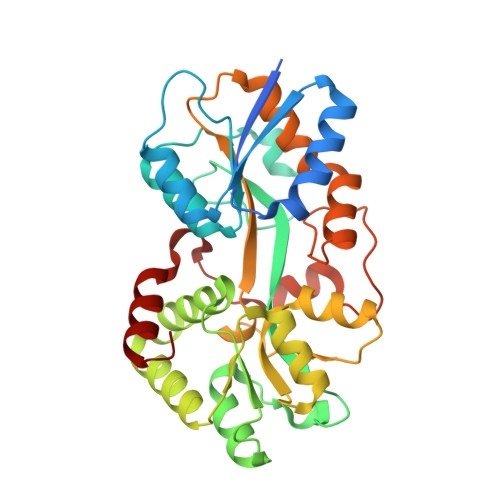
A novel mode of ferric ion coordination by the periplasmic ferric ion-binding subunit FbpA of an ABC-type iron transporter from Thermus thermophilus HB8.
Wang, S., Ogata, M., Horita, S., Ohtsuka, J., Nagata, K., Tanokura, M.(2014) Acta Crystallogr D Biol Crystallogr 70: 196-202
- PubMed: 24419392
- DOI: https://doi.org/10.1107/S1399004713026333
- Primary Citation of Related Structures:
3WAE, 3WAF - PubMed Abstract:
Crystal structures of FbpA, the periplasmic ferric ion-binding protein of an iron-uptake ABC transporter, from Thermus thermophilus HB8 (TtFbpA) have been solved in apo and ferric ion-bound forms at 1.8 and 1.7 Å resolution, respectively. The latter crystal structure shows that the bound ferric ion forms a novel six-coordinated complex with three tyrosine side chains, two bicarbonates and a water molecule in the metal-binding site. The results of gel-filtration chromatography and dynamic light scattering show that TtFbpA exists as a monomer in solution regardless of ferric ion binding and that TtFbpA adopts a more compact conformation in the ferric ion-bound state than in the apo state in solution.
- Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.
Organizational Affiliation: