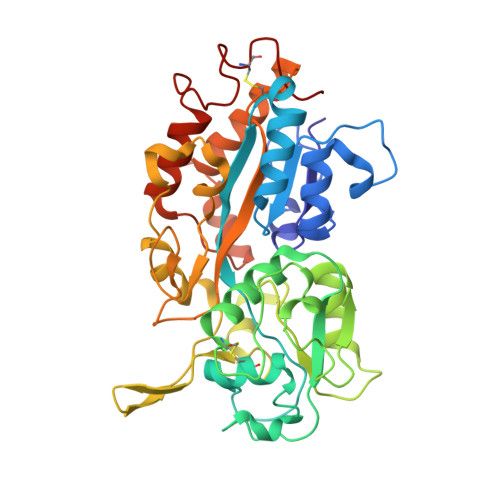
Crystal structure analysis, overexpression and refolding behaviour of a DING protein with single mutation.
Gai, Z.Q., Nakamura, A., Tanaka, Y., Hirano, N., Tanaka, I., Yao, M.(2013) J Synchrotron Radiat 20: 854-858
- PubMed: 24121327
- DOI: https://doi.org/10.1107/S0909049513020694
- Primary Citation of Related Structures:
3W9V, 3W9W - PubMed Abstract:
After crystallization of a certain protein-RNA complex, well diffracting crystals were obtained. However, the asymmetric unit of the crystal was too small to locate any components. Mass spectrometry and X-ray crystal structure analysis showed that it was a member of the DING protein family (HPBP). Surprisingly, the structure of HPBP reported previously was also determined accidentally as a contaminant, suggesting that HPBP has a strong tendency to crystallize. Furthermore, DING proteins were reported to relate in disease. These observations suggest that DING has potential for application in a wide range of research fields. To enable further analyses, a system for preparation of HPBP was constructed. As HPBP was expressed in insoluble form in Escherichia coli, it was unfolded chemically and refolded. Finally, a very high yield preparation method was constructed, in which 43 mg of HPBP was obtained from 1 L of culture. Furthermore, to evaluate the validity of refolding, its crystal structure was determined at 1.03 Å resolution. The determined structure was identical to the native structure, in which two disulfide bonds were recovered correctly and a phosphate ion was captured. Based on these results, it was concluded that the refolded HPBP recovers its structure correctly.
- Faculty of Advanced Life Sciences, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: