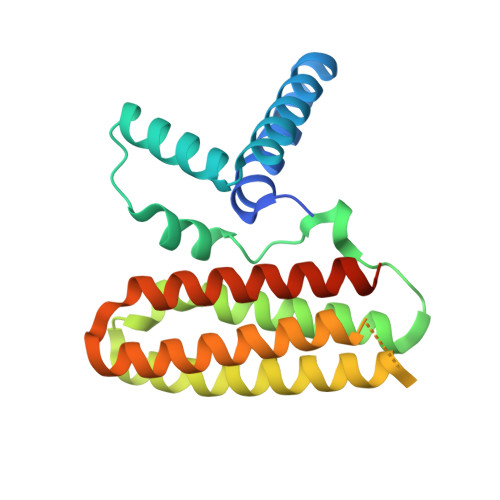
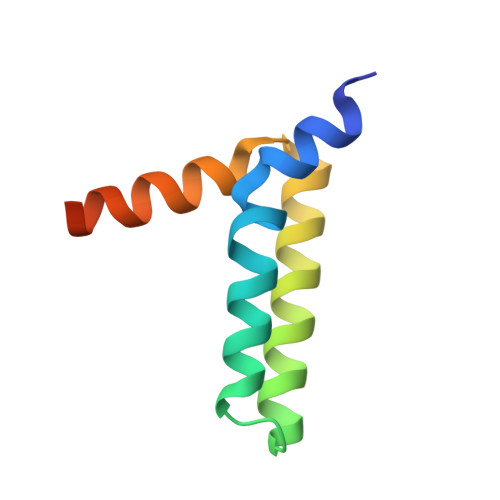
Structural Basis for the Unique Heterodimeric Assembly between Cerebral Cavernous Malformation 3 and Germinal Center Kinase III.
Xu, X., Wang, X., Zhang, Y., Wang, D.C., Ding, J.(2013) Structure 21: 1059-1066
- PubMed: 23665169
- DOI: https://doi.org/10.1016/j.str.2013.04.007
- Primary Citation of Related Structures:
3W8H, 3W8I - PubMed Abstract:
Defects in cerebral cavernous malformation protein CCM3 result in cerebral cavernous malformation (CCM), a common vascular lesion of the human CNS. CCM3 functions as an adaptor protein that interacts with various signal proteins. Among these partner proteins, germinal center kinase III (GCKIII) proteins have attracted significant interest because GCKIII-CCM3 interactions play essential roles in vascular physiology. Here, we report the crystal structures of CCM3 in complex with the C-terminal regulatory domains of GCKIII (GCKIIIct) at 2.4 Å resolution. Our results reveal that GCKIIIct adopts a fold closely resembling that of the CCM3 N-terminal dimeric domain. GCKIIIct heterodimerizes with CCM3 in a manner analogous to CCM3 homodimerization. The remarkable structural rearrangement of CCM3 induced by GCKIIIct binding and the ensuing interactions within CCM3 are characterized as the structural determinants for GCKIIIct-CCM3 heterodimerization. Taken together, these findings provide a precise structural basis for GCKIIIct-CCM3 heterodimerization and the functional performance of GCKIII mediated by CCM3.
- Institute of Biophysics, Chinese Academy of Sciences, National Laboratory of Biomacromolecules, Beijing 100101, People's Republic of China.
Organizational Affiliation: