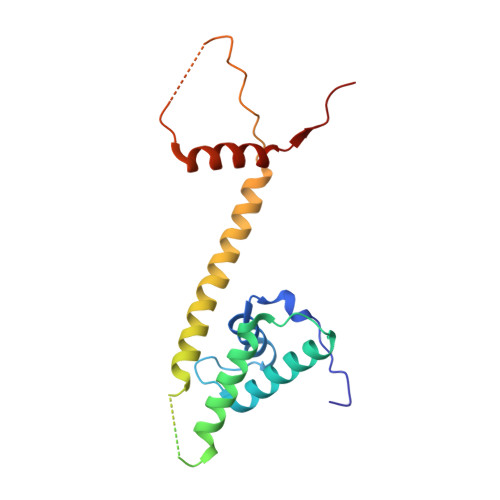
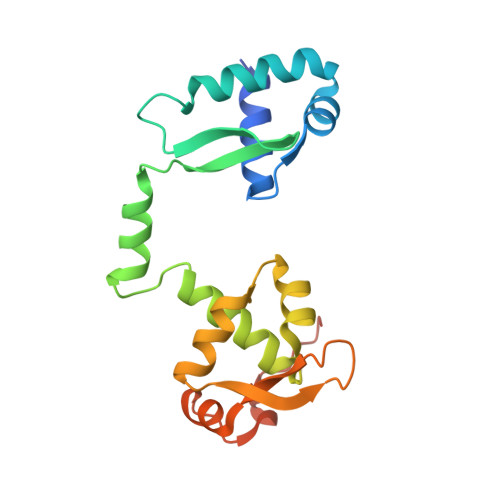
Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB
Kamada, K., Miyata, M., Hirano, T.(2013) Structure 21: 581-594
- PubMed: 23541893
- DOI: https://doi.org/10.1016/j.str.2013.02.016
- Primary Citation of Related Structures:
3W6J, 3W6K - PubMed Abstract:
In many bacteria, a homodimer of structural-maintenance-of-chromosomes proteins associates with two regulatory subunits (known as ScpA and ScpB), assembling a protein complex that plays a crucial role in chromosome organization and segregation. It remains poorly understood, however, how this complex might work at the mechanistic level. Here, we report crystal structures of the ScpAB core complex that display a highly unusual structure in which the central segment of ScpA winds around an asymmetrically oriented ScpB dimer. The two C-terminal domains of the ScpB dimer primarily interact with different regions of ScpA with different affinities. Moreover, flexible interdomain regions of ScpB contribute to a dynamic folding process of the ScpAB subcomplex. Together with other genetic and biochemical assays, we provide evidence that internal structural changes of the ScpAB subcomplex are tightly coupled with activation of the structural-maintenance-of-chromosomes ATPase.
- Chromosome Dynamics Laboratory, RIKEN Advanced Science Institute, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan. kamadak@riken.jp
Organizational Affiliation: