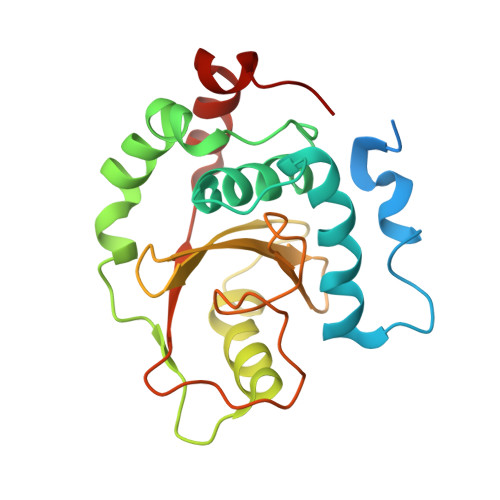
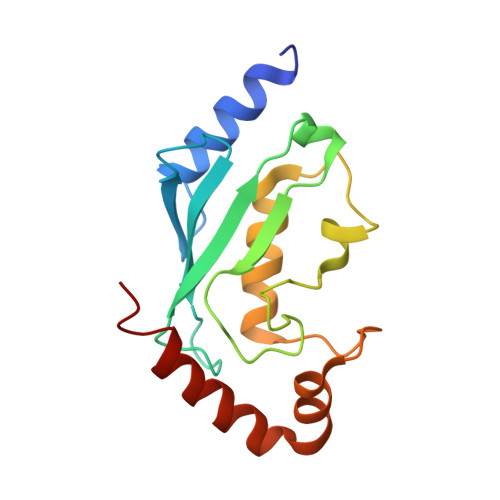
Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
Nishide, A., Kim, M., Takagi, K., Himeno, A., Sanada, T., Sasakawa, C., Mizushima, T.(2013) J Mol Biology 425: 2623-2631
- PubMed: 23542009
- DOI: https://doi.org/10.1016/j.jmb.2013.02.037
- Primary Citation of Related Structures:
3W30, 3W31 - PubMed Abstract:
Ubc13 is a ubiquitin-conjugating enzyme that plays a key role in the nuclear factor-κB signal transduction pathway in human diseases. The Shigella flexneri effector OspI affects inflammatory responses by catalyzing the deamidation of a specific glutamine residue at position 100 in Ubc13 during infection. This modification prevents the activation of the TNF (tumor necrosis factor) receptor-associated factor 6, leading to modulation of the diacylglycerol-CBM (CARD-Bcl10-Malt1) complex-TNF receptor-associated factor 6-nuclear factor-κB signaling pathway. To elucidate the structural basis of OspI function, we determined the crystal structures of the catalytically inert OspI C62A mutant and its complex with Ubc13 at resolutions of 3.0 and 2.96Å, respectively. The structure of the OspI-Ubc13 complex revealed that the interacting surfaces between OspI and Ubc13 are a hydrophobic surface and a complementary charged surface. Furthermore, we predict that the complementary charged surface of OspI plays a key role in substrate specificity determination.
- Picobiology Institute, Department of Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Kouto, Kamigori-cho, Ako-gun, Hyogo 678-1297, Japan.
Organizational Affiliation: