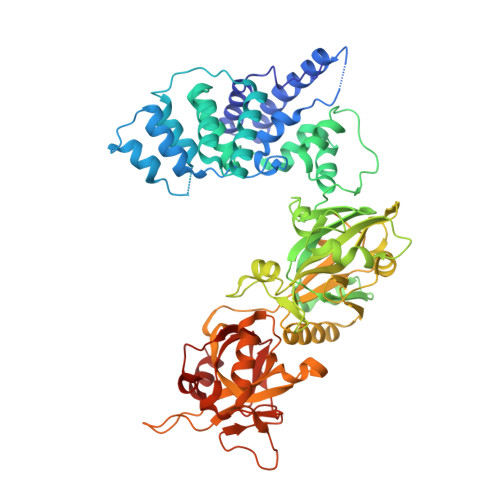
Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Ochi, T., Gu, X., Blundell, T.L.(2013) Structure 21: 672-679
- PubMed: 23523427
- DOI: https://doi.org/10.1016/j.str.2013.02.014
- Primary Citation of Related Structures:
3W1B, 3W1G, 3W5O - PubMed Abstract:
Nonhomologous end joining (NHEJ) is central to the repair of double-stranded DNA breaks throughout the cell cycle and plays roles in the development of the immune system. Although three-dimensional structures of most components of NHEJ have been defined, those of the catalytic region of DNA ligase IV (LigIV), a specialized DNA ligase known to work in NHEJ, and of Artemis have remained unresolved. Here, we report the crystal structure at 2.4 Å resolution of the catalytic region of LigIV (residues 1-609) in complex with an Artemis peptide. We describe interactions of the DNA-binding domain of LigIV with the continuous epitope of Artemis, which, together, form a three-helix bundle. A kink in the first helix of LigIV introduced by a conserved VPF motif gives rise to a hydrophobic pocket, which accommodates a conserved tryptophan from Artemis. We provide structural insights into features of LigIV among human DNA ligases.
- Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK. to237@cam.ac.uk.
Organizational Affiliation: