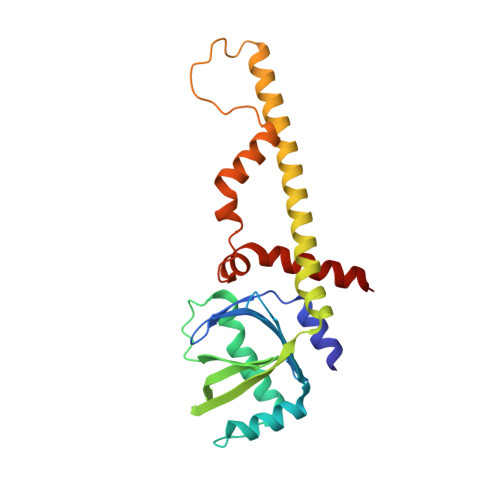
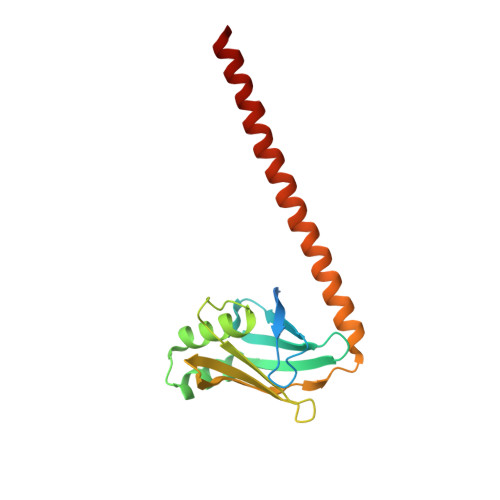
Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Wu, Q., Ochi, T., Matak-Vinkovic, D., Robinson, C.V., Chirgadze, D.Y., Blundell, T.L.(2011) Biochem Soc Trans 39: 1387-1392
- PubMed: 21936820
- DOI: https://doi.org/10.1042/BST0391387
- Primary Citation of Related Structures:
3W03 - PubMed Abstract:
XRCC4 (X-ray cross-complementation group 4) and XLF (XRCC4-like factor) are two essential interacting proteins in the human NHEJ (non-homologous end-joining) pathway that repairs DNA DSBs (double-strand breaks). The individual crystal structures show that the dimeric proteins are homologues with protomers containing head domains and helical coiled-coil tails related by approximate two-fold symmetry. Biochemical, mutagenesis, biophysical and structural studies have identified the regions of interaction between the two proteins and suggested models for the XLF-XRCC4 complex. An 8.5 Å (1 Å = 0.1 nm) resolution crystal structure of XLF-XRCC4 solved by molecular replacement, together with gel filtration and nano-ESI (nano-electrospray ionization)-MS results, demonstrates that XLF and XRCC4 dimers interact through their head domains and form an alternating left-handed helical structure with polypeptide coiled coils and pseudo-dyads of individual XLF and XRCC4 dimers at right angles to the helical axis.
- Department of Biochemistry, University of Cambridge, Tennis Court Road, Cambridge CB2 1GA, UK. qw222@cam.ac.uk
Organizational Affiliation: