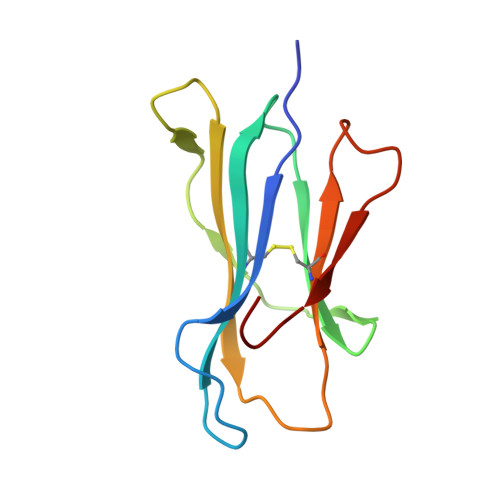
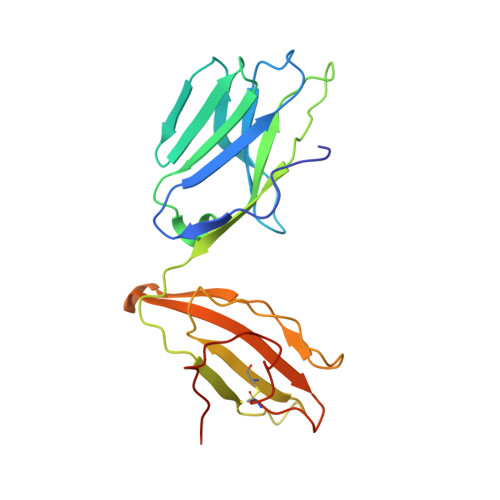
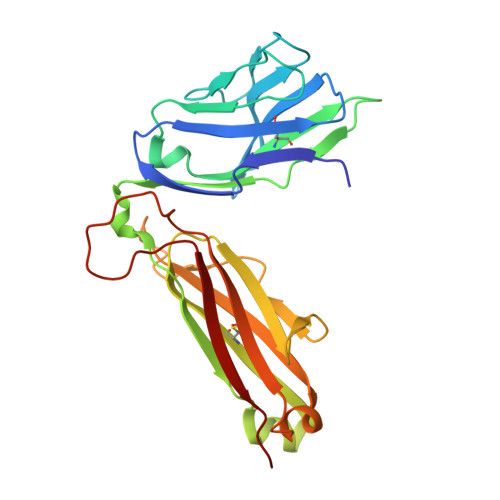
Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
Shimizu, A., Kawana-Tachikawa, A., Yamagata, A., Han, C., Zhu, D., Sato, Y., Nakamura, H., Koibuchi, T., Carlson, J., Martin, E., Brumme, C.J., Shi, Y., Gao, G.F., Brumme, Z.L., Fukai, S., Iwamoto, A.(2013) Sci Rep 3: 3097-3097
- PubMed: 24192765
- DOI: https://doi.org/10.1038/srep03097
- Primary Citation of Related Structures:
3VXM, 3VXN, 3VXO, 3VXP, 3VXQ, 3VXR, 3VXS, 3VXT, 3VXU, 3W0W - PubMed Abstract:
We investigated the crystal structure of an HLA-A*2402-restricted CTL epitope in the HIV-1 nef gene (Nef134-10) before (pHLA) or after TCR docking. The wild type epitope and two escape mutants were included in the study. Y135F was an early-appearing major mutation, while F139L was a late-appearing mutation which was selected in the patients without Y135F. F139 was an eminent feature of the Nef134-10 epitope. Wild type-specific TCR was less fit to F139L mutant suggesting that F139L is an escape from the CTL against the wild type epitope. Although Y135F mutation disrupted the hydrogen bond to HLA-A*2402 His70, newly formed hydrogen bond between T138 and His70 kept the conformation of the epitope in the reconstituted pMHC. TCR from Y135F- or dually-specific CTL had unique mode of binding to the mutant epitope. Y135F has been reported as a processing mutant but CTL carrying structurally adequate TCR can be found in the patients.
- Division of Infectious Diseases, Advanced Clinical Research Center, the Institute of Medical Science, the University of Tokyo. 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan.
Organizational Affiliation: