Crystal Structure of Xylooligosaccharide-Binding Protein from Streptomyces thermoviolaceus OPC-520: Dramatic Conformational Change with Ligand binding
Tomoo, K., Ishida, T., Miyamoto, K., Tsujibo, H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative sugar-binding lipoprotein | 436 | Streptomyces thermoviolaceus | Mutation(s): 0 Gene Names: bxlD | 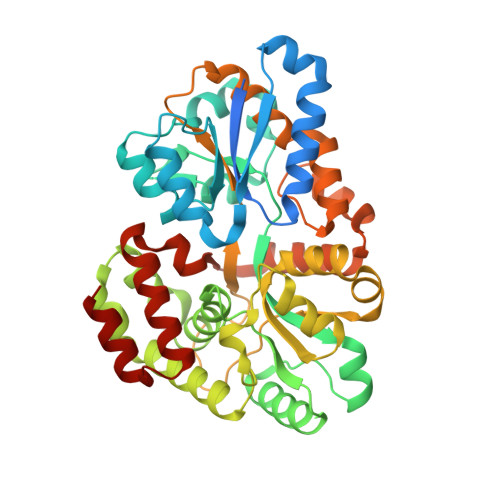 | |
UniProt | |||||
Find proteins for Q76BU9 (Streptomyces thermoviolaceus) Explore Q76BU9 Go to UniProtKB: Q76BU9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q76BU9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900116 Query on PRD_900116 | C, D | 4beta-beta-xylobiose | Oligosaccharide / Metabolism |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.672 | α = 90 |
| b = 163.157 | β = 90 |
| c = 42.931 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |