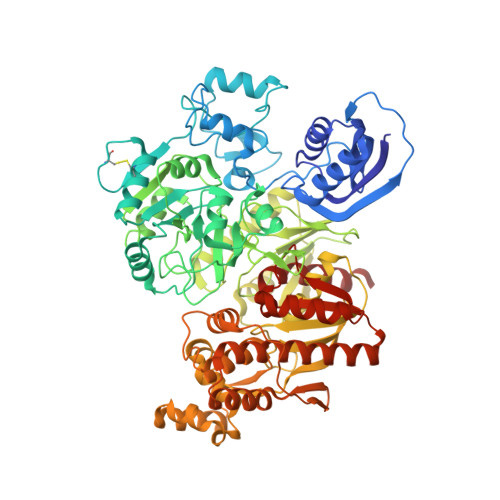
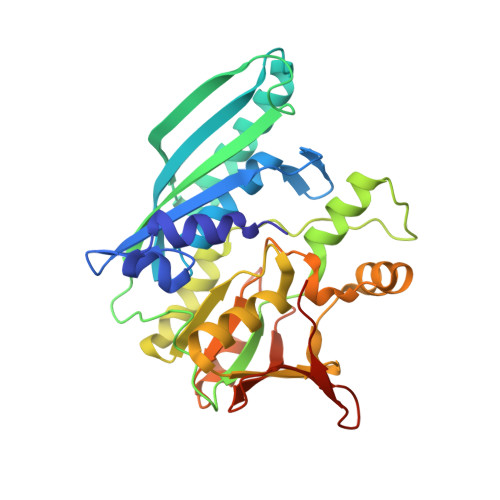
Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases
Shomura, Y., Higuchi, Y.(2012) J Biological Chem 287: 28409-28419
- PubMed: 22740694
- DOI: https://doi.org/10.1074/jbc.M112.387134
- Primary Citation of Related Structures:
3VTH, 3VTI - PubMed Abstract:
As a remarkable structural feature of hydrogenase active sites, [NiFe]-hydrogenases harbor one carbonyl and two cyano ligands, where HypE and HypF are involved in the biosynthesis of the nitrile group as a precursor of the cyano groups. HypF catalyzes S-carbamoylation of the C-terminal cysteine of HypE via three steps using carbamoylphosphate and ATP, producing two unstable intermediates: carbamate and carbamoyladenylate. Although the crystal structures of intact HypE homodimers and partial HypF have been reported, it remains unclear how the consecutive reactions occur without the loss of unstable intermediates during the proposed reaction scheme. Here we report the crystal structures of full-length HypF both alone and in complex with HypE at resolutions of 2.0 and 2.6 Å, respectively. Three catalytic sites of the structures of the HypF nucleotide- and phosphate-bound forms have been identified, with each site connected via channels inside the protein. This finding suggests that the first two consecutive reactions occur without the release of carbamate or carbamoyladenylate from the enzyme. The structure of HypF in complex with HypE revealed that HypF can associate with HypE without disturbing its homodimeric interaction and that the binding manner allows the C-terminal Cys-351 of HypE to access the S-carbamoylation active site in HypF, suggesting that the third step can also proceed without the release of carbamoyladenylate. A comparison of the structure of HypF with the recently reported structures of O-carbamoyltransferase revealed different reaction mechanisms for carbamoyladenylate synthesis and a similar reaction mechanism for carbamoyltransfer to produce the carbamoyl-HypE molecule.
- Department of Life Science, Graduate School of Life Science, University of Hyogo, Hyogo 678-1297, Japan. shomura@sci.u-hyogo.ac.jp
Organizational Affiliation: