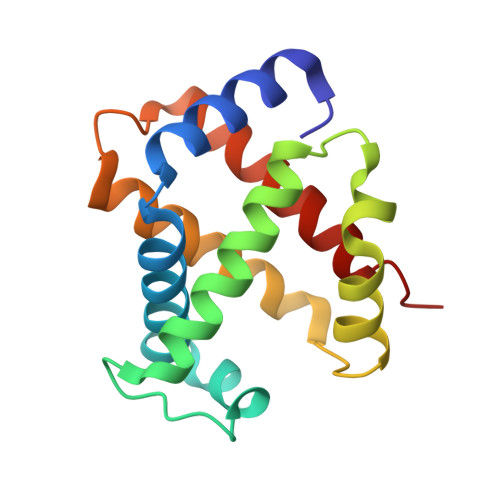
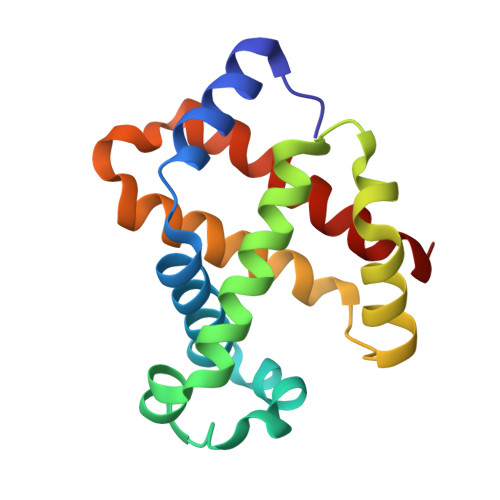
Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Noguchi, H., Campbell, K.L., Ho, C., Unzai, S., Park, S.-Y., Tame, J.R.H.(2012) Acta Crystallogr D Biol Crystallogr 68: 1441-1449
- PubMed: 23090393
- DOI: https://doi.org/10.1107/S0907444912029459
- Primary Citation of Related Structures:
3VRE, 3VRF, 3VRG - PubMed Abstract:
The haemoglobin (Hb) of the extinct woolly mammoth has been recreated using recombinant genes expressed in Escherichia coli. The globin gene sequences were previously determined using DNA recovered from frozen cadavers. Although highly similar to the Hb of existing elephants, the woolly mammoth protein shows rather different responses to chloride ions and temperature. In particular, the heat of oxygenation is found to be much lower in mammoth Hb, which appears to be an adaptation to the harsh high-latitude climates of the Pleistocene Ice Ages and has been linked to heightened sensitivity of the mammoth protein to protons, chloride ions and organic phosphates relative to that of Asian elephants. To elucidate the structural basis for the altered homotropic and heterotropic effects, the crystal structures of mammoth Hb have been determined in the deoxy, carbonmonoxy and aquo-met forms. These models, which are the first structures of Hb from an extinct species, show many features reminiscent of human Hb, but underline how the delicate control of oxygen affinity relies on much more than simple overall quaternary-structure changes.
- Protein Design Laboratory, Yokohama City University, Suehiro 1-7-29, Yokohama 230-0045, Japan.
Organizational Affiliation: