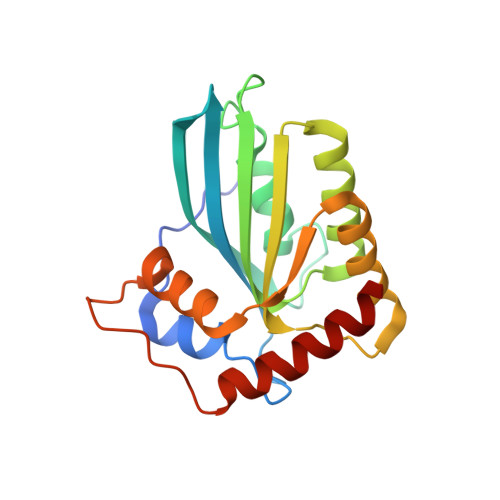
The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Kitamura, S., Ode, H., Nakashima, M., Imahashi, M., Naganawa, Y., Kurosawa, T., Yokomaku, Y., Yamane, T., Watanabe, N., Suzuki, A., Sugiura, W., Iwatani, Y.(2012) Nat Struct Mol Biol 19: 1005-1010
- PubMed: 23001005
- DOI: https://doi.org/10.1038/nsmb.2378
- Primary Citation of Related Structures:
3VOW - PubMed Abstract:
The human apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3, referred to as A3) proteins are cellular cytidine deaminases that potently restrict retrovirus replication. However, HIV-1 viral infectivity factor (Vif) counteracts the antiviral activity of most A3 proteins by targeting them for proteasomal degradation. To date, the structure of an A3 protein containing a Vif-binding interface has not been solved. Here, we report a high-resolution crystal structure of APOBEC3C and identify the HIV-1 Vif-interaction interface. Extensive structure-guided mutagenesis revealed the role of a shallow cavity composed of hydrophobic or negatively charged residues between the α2 and α3 helices. This region is distant from the DPD motif (residues 128-130) of APOBEC3G that participates in HIV-1 Vif interaction. These findings provide insight into Vif-A3 interactions and could lead to the development of new pharmacologic anti-HIV-1 compounds.
- Clinical Research Center, Department of Infectious Diseases and Immunology, National Hospital Organization Nagoya Medical Center, Nagoya, Japan.
Organizational Affiliation: