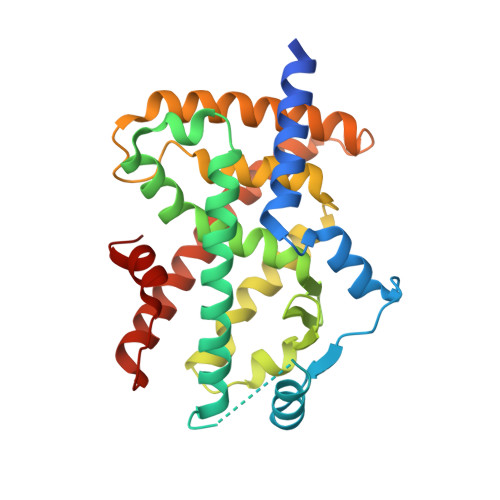
Structural basis for telmisartan-mediated partial activation of PPAR gamma
Amano, Y., Yamaguchi, T., Ohno, K., Niimi, T., Orita, M., Sakashita, H., Takeuchi, M.(2012) Hypertens Res 35: 715-719
- PubMed: 22357520
- DOI: https://doi.org/10.1038/hr.2012.17
- Primary Citation of Related Structures:
3VN2 - PubMed Abstract:
Telmisartan, a selective angiotensin II type 1 receptor blocker, has recently been shown to act as a partial agonist for peroxisome proliferator-activated receptor gamma (PPARγ). To understand how telmisartan partially activates PPARγ, we determined the ternary complex structure of PPARγ, telmisartan, and a coactivator peptide from steroid receptor coactivator-1 at a resolution of 2.18 Å. Crystallographic analysis revealed that telmisartan exhibits an unexpected binding mode in which the central benzimidazole ring is engaged in a non-canonical--and suboptimal--hydrogen-bonding network around helix 12 (H12). This network differs greatly from that observed when full-agonists bind with PPARγ and prompt high-coactivator recruitment through H12 stabilized by multiple hydrogen bonds. Binding with telmisartan results in a less stable H12 that in turn leads to attenuated coactivator binding, thus explaining the mechanism of partial activation.
- Drug Discovery Research, Astellas Pharma Inc., Ibaraki, Japan.
Organizational Affiliation: