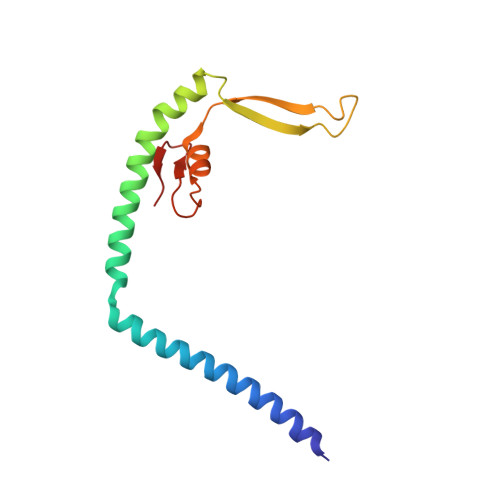
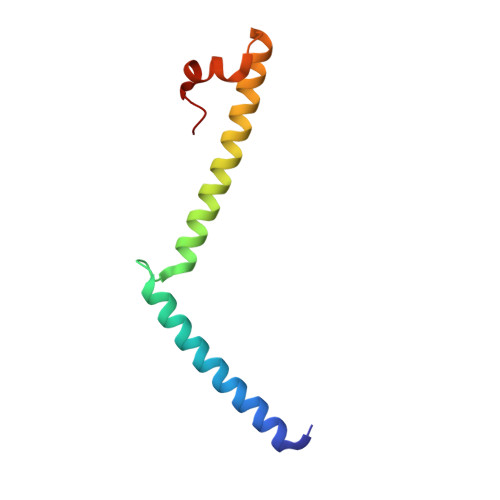
Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Kuwabara, N., Murayama, Y., Hashimoto, H., Kokabu, Y., Ikeguchi, M., Sato, M., Mayanagi, K., Tsutsui, Y., Iwasaki, H., Shimizu, T.(2012) Structure 20: 440-449
- PubMed: 22405003
- DOI: https://doi.org/10.1016/j.str.2012.01.005
- Primary Citation of Related Structures:
3VIQ, 3VIR - PubMed Abstract:
Rad51 forms a helical filament on single-stranded DNA and promotes strand exchange between two homologous DNA molecules during homologous recombination. The Swi5-Sfr1 complex interacts directly with Rad51 and stimulates strand exchange. Here we describe structural and functional aspects of the complex. Swi5 and the C-terminal core domain of Sfr1 form an essential activator complex with a parallel coiled-coil heterodimer joined firmly together via two previously uncharacterized leucine-zipper motifs and a bundle. The resultant coiled coil is sharply kinked, generating an elongated crescent-shaped structure suitable for transient binding within the helical groove of the Rad51 filament. The N-terminal region of Sfr1, meanwhile, has an interface for binding of Rad51. Our data suggest that the snug fit resulting from the complementary geometry of the heterodimer activates the Rad51 filament and that the N-terminal domain of Sfr1 plays a role in the efficient recruitment of the Swi5-Sfr1 complex to the Rad51 filaments.
- Graduate School of Pharmaceutical Sciences, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: