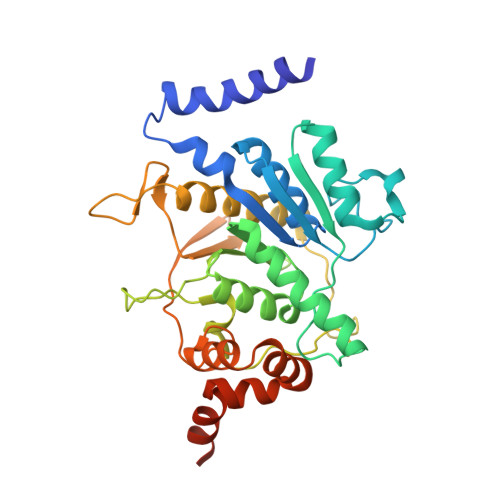
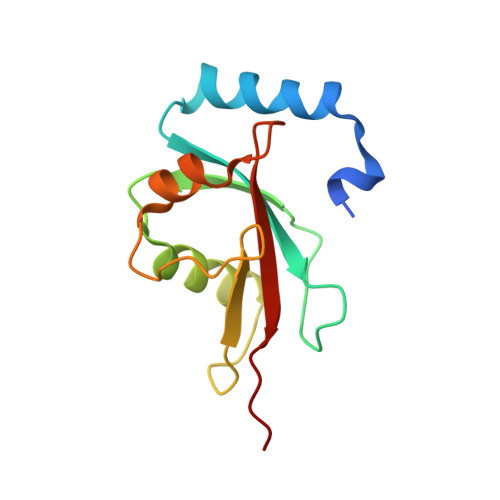
Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Noda, N.N., Satoo, K., Fujioka, Y., Kumeta, H., Ogura, K., Nakatogawa, H., Ohsumi, Y., Inagaki, F.(2011) Mol Cell 44: 462-475
- PubMed: 22055191
- DOI: https://doi.org/10.1016/j.molcel.2011.08.035
- Primary Citation of Related Structures:
2LI5, 3VH1, 3VH2, 3VH3, 3VH4 - PubMed Abstract:
E1 enzymes activate ubiquitin-like proteins and transfer them to cognate E2 enzymes. Atg7, a noncanonical E1, activates two ubiquitin-like proteins, Atg8 and Atg12, and plays a crucial role in autophagy. Here, we report crystal structures of full-length Atg7 and its C-terminal domain bound to Atg8 and MgATP, as well as a solution structure of Atg8 bound to the extreme C-terminal domain (ECTD) of Atg7. The unique N-terminal domain (NTD) of Atg7 is responsible for Atg3 (E2) binding, whereas its C-terminal domain is comprised of a homodimeric adenylation domain (AD) and ECTD. The structural and biochemical data demonstrate that Atg8 is initially recognized by the C-terminal tail of ECTD and is then transferred to an AD, where the Atg8 C terminus is attacked by the catalytic cysteine to form a thioester bond. Atg8 is then transferred via a trans mechanism to the Atg3 bound to the NTD of the opposite protomer within a dimer.
- Institute of Microbial Chemistry, Tokyo, Tokyo 141-0021, Japan. nn@bikaken.or.jp
Organizational Affiliation: