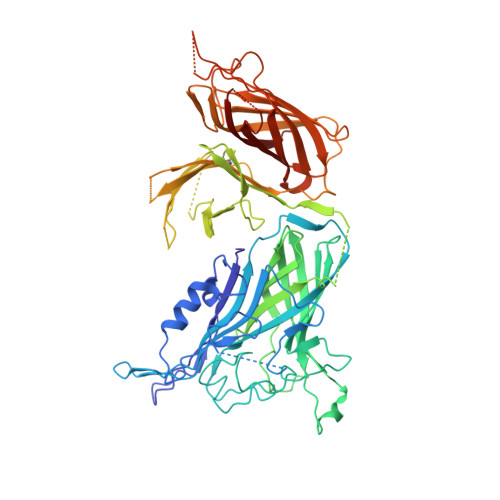
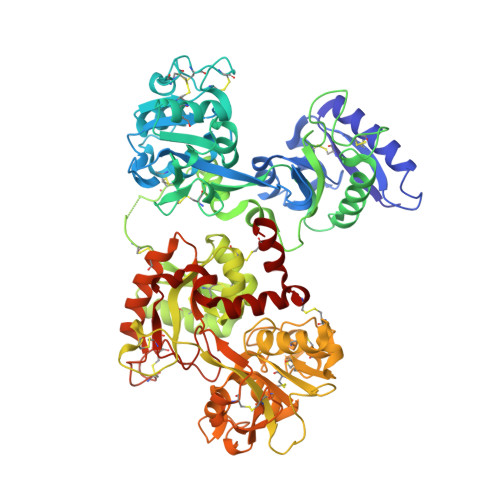
The structural basis of transferrin sequestration by transferrin-binding protein B.
Calmettes, C., Alcantara, J., Yu, R.H., Schryvers, A.B., Moraes, T.F.(2012) Nat Struct Mol Biol 19: 358-360
- PubMed: 22343719
- DOI: https://doi.org/10.1038/nsmb.2251
- Primary Citation of Related Structures:
3VE1, 3VE2 - PubMed Abstract:
Neisseria meningitidis, the causative agent of bacterial meningitis, acquires the essential element iron from the host glycoprotein transferrin during infection through a surface transferrin receptor system composed of proteins TbpA and TbpB. Here we present the crystal structures of TbpB from N. meningitidis in its apo form and in complex with human transferrin. The structure reveals how TbpB sequesters and initiates iron release from human transferrin.
- Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: