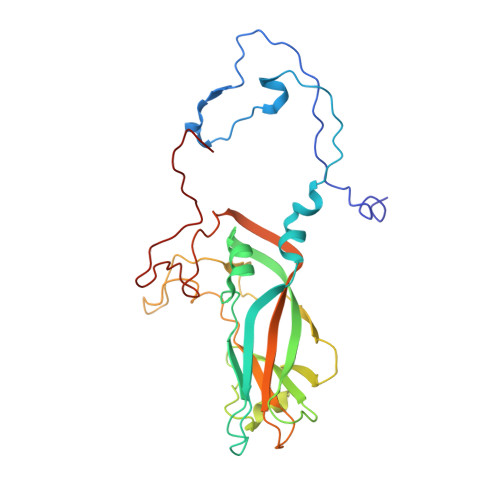
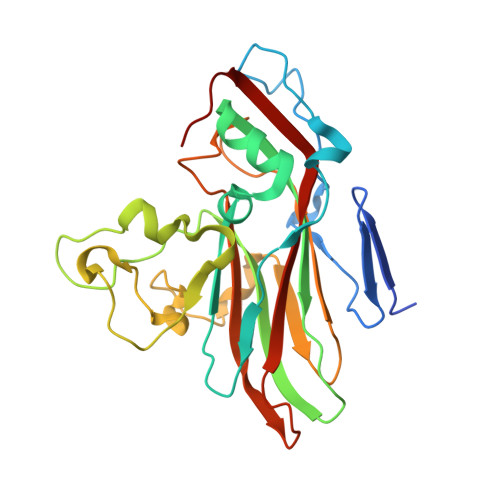
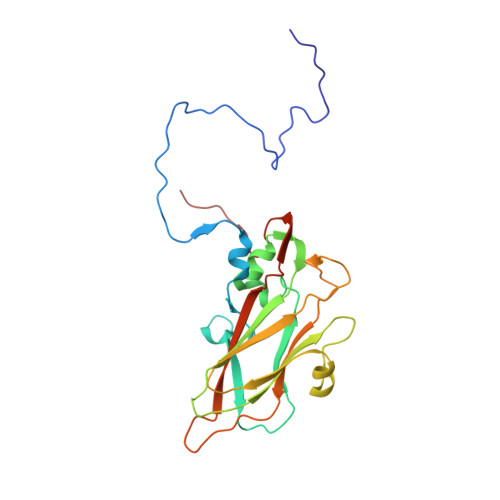
An Orally Available 3-Ethoxybenzisoxazole Capsid Binder with Clinical Activity against Human Rhinovirus.
Feil, S.C., Hamilton, S., Krippner, G.Y., Lin, B., Luttick, A., McConnell, D.B., Nearn, R., Parker, M.W., Ryan, J., Stanislawski, P.C., Tucker, S.P., Watson, K.G., Morton, C.J.(2012) ACS Med Chem Lett 3: 303-307
- PubMed: 24900468
- DOI: https://doi.org/10.1021/ml2002955
- Primary Citation of Related Structures:
3VDD - PubMed Abstract:
Respiratory infections caused by human rhinovirus are responsible for severe exacerbations of underlying clinical conditions such as asthma in addition to their economic cost in terms of lost working days due to illness. While several antiviral compounds for treating rhinoviral infections have been discovered, none have succeeded, to date, in reaching approval for clinical use. We have developed a potent, orally available rhinovirus inhibitor 6 that has progressed through early clinical trials. The compound shows favorable pharmacokinetic and activity profiles and has a confirmed mechanism of action through crystallographic studies of a rhinovirus-compound complex. The compound has now progressed to phase IIb clinical studies of its effect on natural rhinovirus infection in humans.
- Biota Structural Biology Laboratory, St. Vincent's Institute , 9 Princes Street, Fitzroy, Victoria 3065, Australia.
Organizational Affiliation: