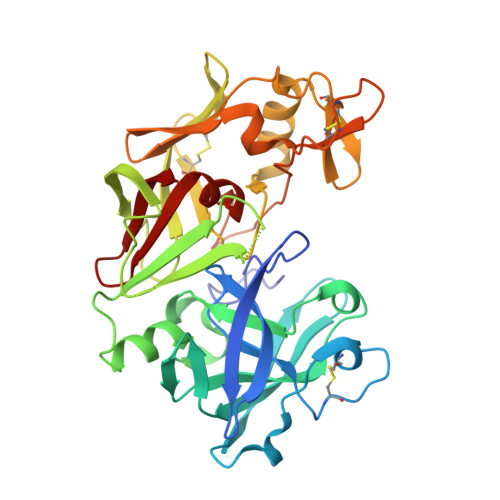
Human Prorenin Structure Sheds Light on a Novel Mechanism of Its Autoinhibition and on Its Non-Proteolytic Activation by the (Pro)renin Receptor.
Morales, R., Watier, Y., Bocskei, Z.(2012) J Mol Biology 421: 100-111
- PubMed: 22575890
- DOI: https://doi.org/10.1016/j.jmb.2012.05.003
- Primary Citation of Related Structures:
3VCM - PubMed Abstract:
Antibodies and prorenin mutants have long been used to structurally characterize prorenin, the inactive proenzyme form of renin. They were designed on the basis of homology models built using other aspartyl protease proenzyme structures since no structure was available for prorenin. Here, we present the first X-ray structure of a prorenin. The current structure of prorenin reveals that, in this zymogene, the active site of renin is blocked by the N-terminal residues of the mature version of the renin molecule, which are, in turn, covered by an Ω-shaped prosegment. This prevents access of substrates to the active site. The departure of the prosegment on activation induces an important global conformational change in the mature renin molecule with respect to prorenin: similar to other related enzymes such as pepsin or gastricsin, the segment that constitutes the N-terminal β-strand in renin is displaced from the renin active site by about 180° straight into the position that corresponds to the N-terminal β-strand of the prorenin prosegment. This way, the renin active site will become completely exposed and capable of carrying out its catalytic functions. A unique inactivation mechanism is also revealed, which does not make use of a lysine against the catalytic aspartates, probably in order to facilitate pH-independent activation [e.g., by the (pro)renin receptor].
- Sanofi-Aventis R&D, LGCR Structure Design and Informatics, 16 Rue d'Ankara, 67000 Strasbourg, France.
Organizational Affiliation: