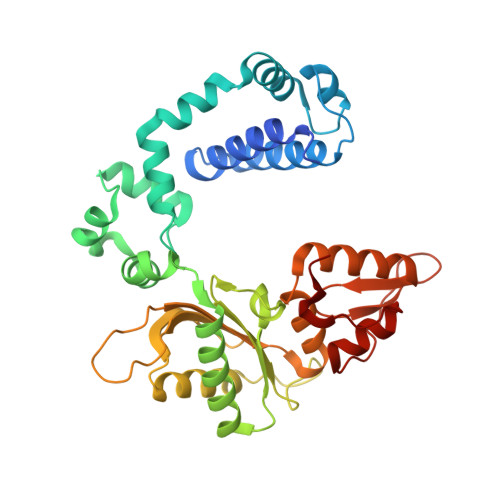
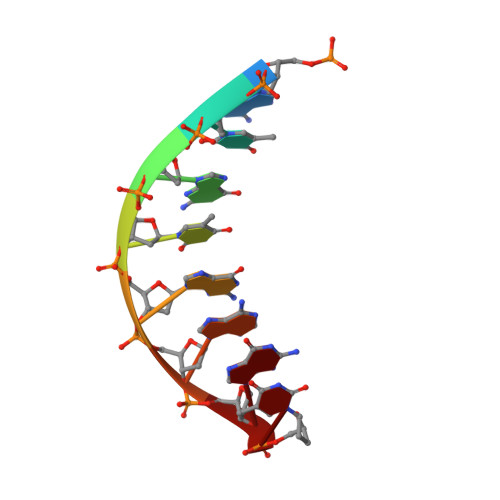
Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
Rangarajan, S., Gridley, C.L., Firbank, S., Dalal, S., Sweasy, J.B., Jaeger, J.To be published.