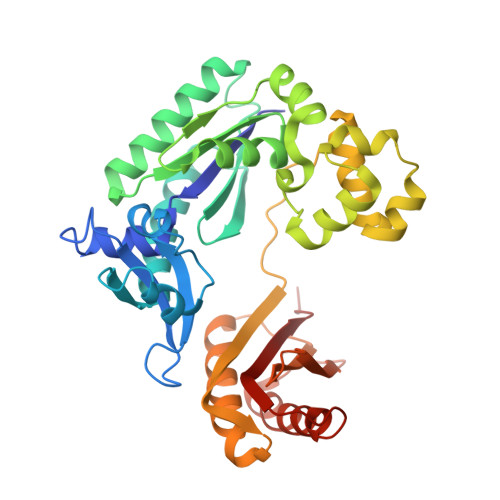
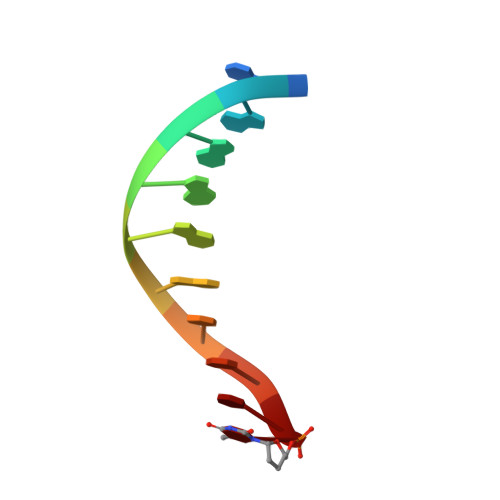
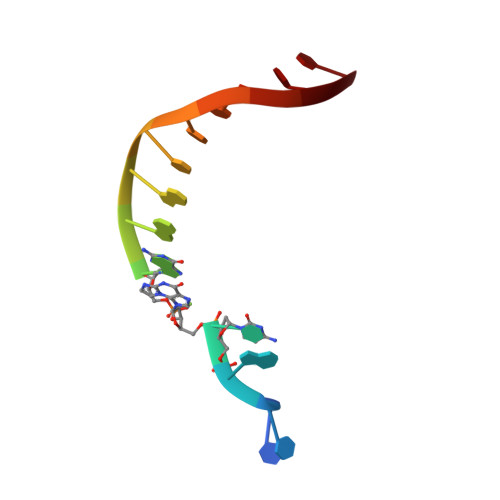
Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Zhao, L., Christov, P.P., Kozekov, I.D., Pence, M.G., Pallan, P.S., Rizzo, C.J., Egli, M., Guengerich, F.P.(2012) Angew Chem Int Ed Engl 51: 5466-5469
- PubMed: 22488769
- DOI: https://doi.org/10.1002/anie.201109004
- Primary Citation of Related Structures:
3V6H, 3V6J, 3V6K - Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, TN 37232-0146, USA.
Organizational Affiliation: