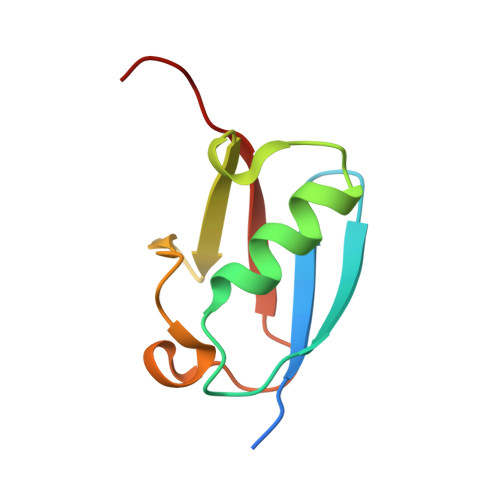
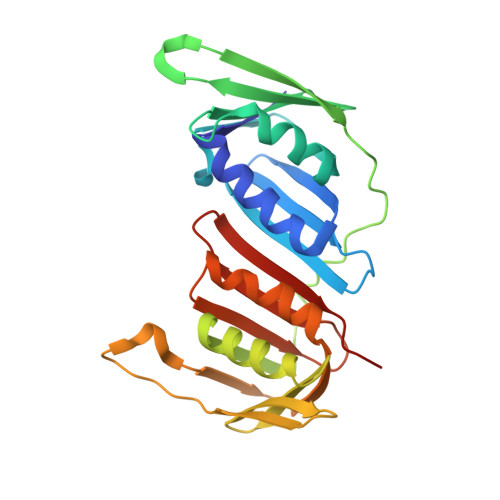
Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2.
Armstrong, A.A., Mohideen, F., Lima, C.D.(2012) Nature 483: 59-63
- PubMed: 22382979
- DOI: https://doi.org/10.1038/nature10883
- Primary Citation of Related Structures:
3V60, 3V61, 3V62 - PubMed Abstract:
Ubiquitin (Ub) and ubiquitin-like (Ubl) modifiers such as SUMO (also known as Smt3 in Saccharomyces cerevisiae) mediate signal transduction through post-translational modification of substrate proteins in pathways that control differentiation, apoptosis and the cell cycle, and responses to stress such as the DNA damage response. In yeast, the proliferating cell nuclear antigen PCNA (also known as Pol30) is modified by ubiquitin in response to DNA damage and by SUMO during S phase. Whereas Ub-PCNA can signal for recruitment of translesion DNA polymerases, SUMO-PCNA signals for recruitment of the anti-recombinogenic DNA helicase Srs2. It remains unclear how receptors such as Srs2 specifically recognize substrates after conjugation to Ub and Ubls. Here we show, through structural, biochemical and functional studies, that the Srs2 carboxy-terminal domain harbours tandem receptor motifs that interact independently with PCNA and SUMO and that both motifs are required to recognize SUMO-PCNA specifically. The mechanism presented is pertinent to understanding how other receptors specifically recognize Ub- and Ubl-modified substrates to facilitate signal transduction.
- Structural Biology Program, Sloan-Kettering Institute, New York, New York 10065, USA.
Organizational Affiliation: