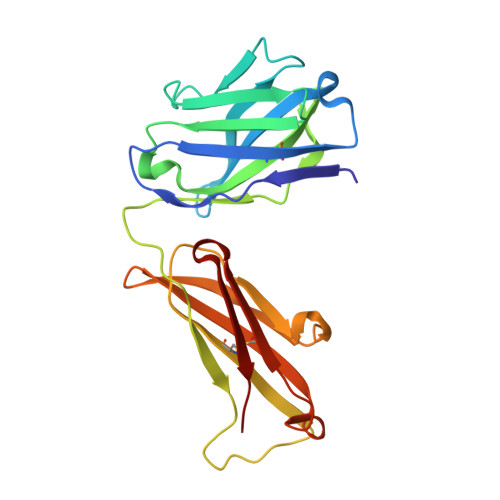
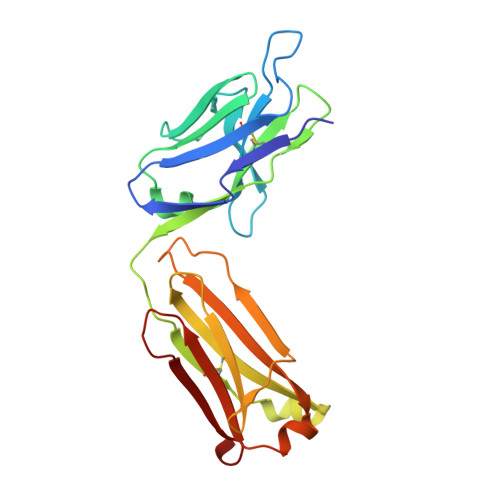
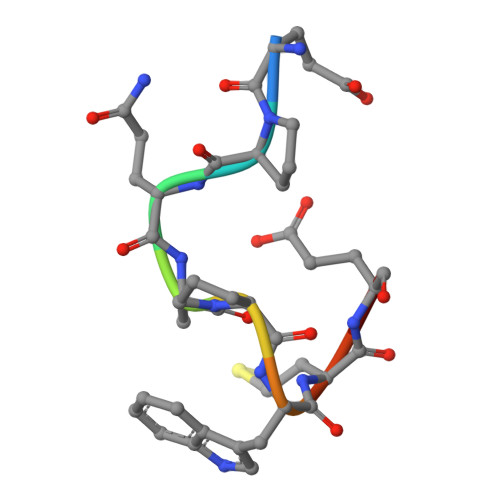
The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
Mage, M.G., Dolan, M.A., Wang, R., Boyd, L.F., Revilleza, M.J., Robinson, H., Natarajan, K., Myers, N.B., Hansen, T.H., Margulies, D.H.(2012) J Immunol 189: 1391-1399
- PubMed: 22753930
- DOI: https://doi.org/10.4049/jimmunol.1200831
- Primary Citation of Related Structures:
3UO1, 3UYR, 3V4U, 3V52 - PubMed Abstract:
MHC class I (MHC-I) proteins of the adaptive immune system require antigenic peptides for maintenance of mature conformation and immune function via specific recognition by MHC-I-restricted CD8(+) T lymphocytes. New MHC-I molecules in the endoplasmic reticulum are held by chaperones in a peptide-receptive (PR) transition state pending release by tightly binding peptides. In this study, we show, by crystallographic, docking, and molecular dynamics methods, dramatic movement of a hinged unit containing a conserved 3(10) helix that flips from an exposed "open" position in the PR transition state to a "closed" position with buried hydrophobic side chains in the peptide-loaded mature molecule. Crystallography of hinged unit residues 46-53 of murine H-2L(d) MHC-I H chain, complexed with mAb 64-3-7, demonstrates solvent exposure of these residues in the PR conformation. Docking and molecular dynamics predict how this segment moves to help form the A and B pockets crucial for the tight peptide binding needed for stability of the mature peptide-loaded conformation, chaperone dissociation, and Ag presentation.
- Molecular Biology Section, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA. mmage@mail.nih.gov
Organizational Affiliation: