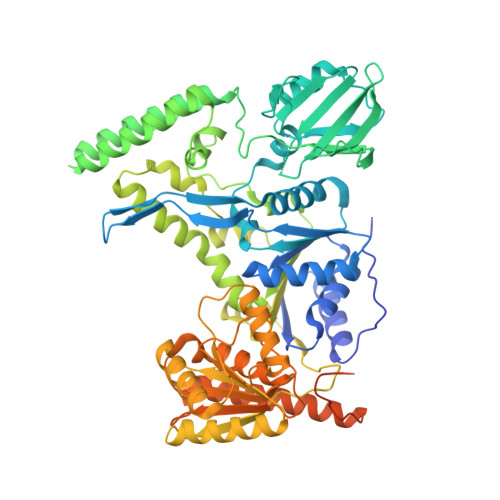
Crystal structure of the UvrB dimer: insights into the nature and functioning of the UvrAB damage engagement and UvrB-DNA complexes.
Webster, M.P., Jukes, R., Zamfir, V.S., Kay, C.W., Bagneris, C., Barrett, T.(2012) Nucleic Acids Res 40: 8743-8758
- PubMed: 22753105
- DOI: https://doi.org/10.1093/nar/gks633
- Primary Citation of Related Structures:
3V4R - PubMed Abstract:
UvrB has a central role in the highly conserved UvrABC pathway functioning not only as a damage recognition element but also as an essential component of the lesion tracking machinery. While it has been recently confirmed that the tracking assembly comprises a UvrA2B2 heterotetramer, the configurations of the damage engagement and UvrB-DNA handover complexes remain obscure. Here, we present the first crystal structure of a UvrB dimer whose biological significance has been verified using both chemical cross-linking and electron paramagnetic resonance spectroscopy. We demonstrate that this dimeric species stably associates with UvrA and forms a UvrA2B2-DNA complex. Our studies also illustrate how signals are transduced between the ATP and DNA binding sites to generate the helicase activity pivotal to handover and formation of the UvrB2-DNA complex, providing key insights into the configurations of these important repair intermediates.
- Department of Biological Sciences, Institute of Structural and Molecular Biology, Crystallography, Birkbeck College, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: