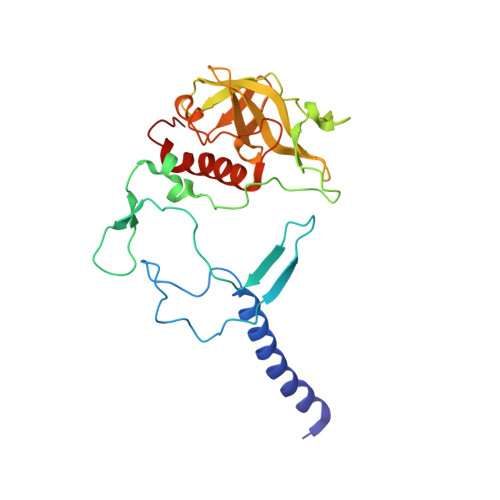
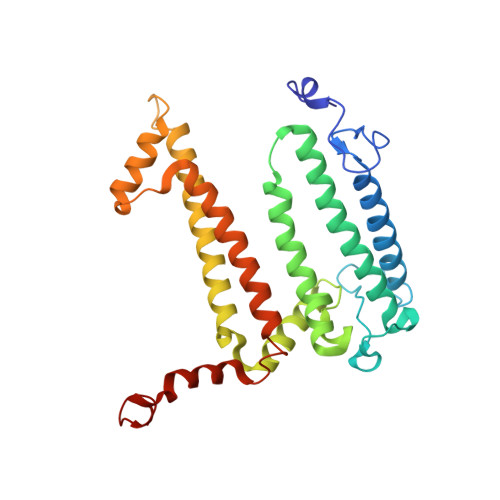
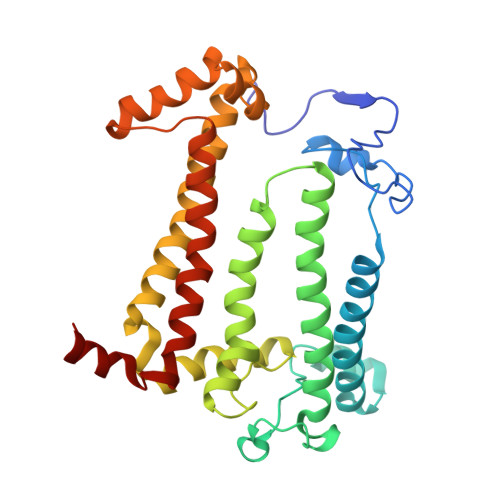
The site-directed mutation I(L177)H in Rhodobacter sphaeroides reaction center affects coordination of P(A) and B(B) bacteriochlorophylls.
Vasilieva, L.G., Fufina, T.Y., Gabdulkhakov, A.G., Leonova, M.M., Khatypov, R.A., Shuvalov, V.A.(2012) Biochim Biophys Acta 1817: 1407-1417
- PubMed: 22365928
- DOI: https://doi.org/10.1016/j.bbabio.2012.02.008
- Primary Citation of Related Structures:
3V3Y, 3V3Z - PubMed Abstract:
To explore the influence of the I(L177)H single mutation on the properties of the nearest bacteriochlorophylls (BChls), three reaction centers (RCs) bearing double mutations were constructed in the photosynthetic purple bacterium Rhodobacter sphaeroides, and their properties and pigment content were compared with those of the correspondent single mutant RCs. Each pair of the mutations comprised the amino acid substitution I(L177)H and another mutation altering histidine ligand of BChl P(A) or BChl B(B). Contrary to expectations, the double mutation I(L177)H+H(L173)L does not bring about a heterodimer RC but causes a 46nm blue shift of the long-wavelength P absorbance band. The histidine L177 or a water molecule were suggested as putative ligands for P(A) in the RC I(L177)H+H(L173)L although this would imply a reorientation of the His backbone and additional rearrangements in the primary donor environment or even a repositioning of the BChl dimer. The crystal structure of the mutant I(L177)H reaction center determined to a resolution of 2.9Å shows changes at the interface region between the BChl P(A) and the monomeric BChl B(B). Spectral and pigment analysis provided evidence for β-coordination of the BChl B(B) in the double mutant RC I(L177)H+H(M182)L and for its hexacoordination in the mutant reaction center I(L177)H. Computer modeling suggests involvement of two water molecules in the β-coordination of the BChl B(B). Possible structural consequences of the L177 mutation affecting the coordination of the two BChls P(A) and B(B) are discussed. This article is part of a Special Issue entitled: Photosynthesis Research for Sustainability: from Natural to Artificial.
- Institute of Basic Biological Problems, Russian Academy of Sciences, Pushchino, Moscow, Russia.
Organizational Affiliation: