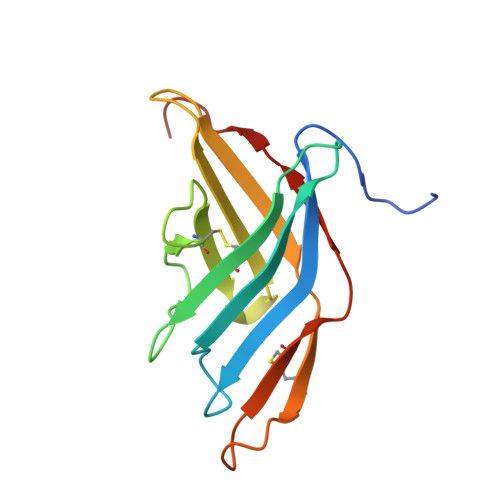
Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
Chruszcz, M., Chapman, M.D., Osinski, T., Solberg, R., Demas, M., Porebski, P.J., Majorek, K.A., Pomes, A., Minor, W.(2012) J Allergy Clin Immunol 130: 241-7.e9
- PubMed: 22664167
- DOI: https://doi.org/10.1016/j.jaci.2012.03.047
- Primary Citation of Related Structures:
3V0R - PubMed Abstract:
Alternaria species is one of the most common molds associated with allergic diseases, and 80% of Alternaria species-sensitive patients produce IgE antibodies to a major protein allergen, Alt a 1. The structure and function of Alt a 1 is unknown. We sought to obtain a high-resolution structure of Alt a 1 using x-ray crystallography and to investigate structural relationships between Alt a 1 and other allergens and proteins reported in the Protein Data Bank. X-ray crystallography was used to determine the structure of Alt a 1 by using a custom-designed set of crystallization conditions. An initial Alt a 1 model was determined by the application of a Ta(6)Br(12)(2+) cluster and single-wavelength anomalous diffraction. Bioinformatic analyses were used to compare the Alt a 1 sequence and structure with that of other proteins. Alt a 1 is a unique β-barrel comprising 11 β-strands and forms a "butterfly-like" dimer linked by a single disulfide bond with a large (1345 Å(2)) dimer interface. Intramolecular disulfide bonds are conserved among Alt a 1 homologs. Currently, the Alt a 1 structure has no equivalent in the Protein Data Bank. Bioinformatics analyses suggest that the structure is found exclusively in fungi. Four previously reported putative IgE-binding peptides have been located on the Alt a 1 structure. Alt a 1 has a unique, dimeric β-barrel structure that appears to define a new protein family with unknown function found exclusively in fungi. The location of IgE antibody-binding epitopes is in agreement with the structural analysis of Alt a 1. The Alt a 1 structure will allow mechanistic structure/function studies and immunologic studies directed toward new forms of immunotherapy for Alternaria species-sensitive allergic patients.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA. maks@iwonka.med.virginia.edu
Organizational Affiliation: