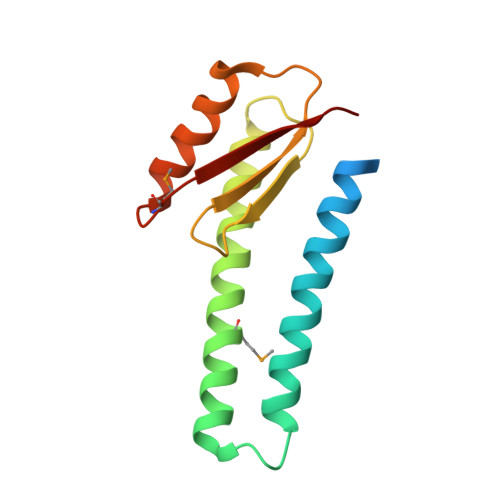
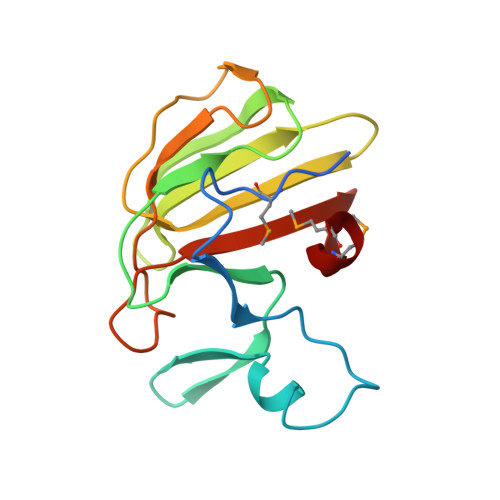
Structure of the basal components of a bacterial transporter.
Meisner, J., Maehigashi, T., Andre, I., Dunham, C.M., Moran, C.P.(2012) Proc Natl Acad Sci U S A 109: 5446-5451
- PubMed: 22431613
- DOI: https://doi.org/10.1073/pnas.1120113109
- Primary Citation of Related Structures:
3UZ0 - PubMed Abstract:
Proteins SpoIIQ and SpoIIIAH interact through two membranes to connect the forespore and the mother cell during endospore development in the bacterium Bacillus subtilis. SpoIIIAH consists of a transmembrane segment and an extracellular domain with similarity to YscJ proteins. YscJ proteins form large multimeric rings that are the structural scaffolds for the assembly of type III secretion systems in gram-negative bacteria. The predicted ring-forming motif of SpoIIIAH and other evidence led to the model that SpoIIQ and SpoIIIAH form the core components of a channel or transporter through which the mother cell nurtures forespore development. Therefore, to understand the roles of SpoIIIAH and SpoIIQ in channel formation, it is critical to determine whether SpoIIIAH adopts a ring-forming structural motif, and whether interaction of SpoIIIAH with SpoIIQ would preclude ring formation. We report a 2.8-Å resolution structure of a complex of SpoIIQ and SpoIIIAH. SpoIIIAH folds into the ring-building structural motif, and modeling shows that the structure of the SpoIIQ-SpoIIIAH complex is compatible with forming a symmetrical oligomer that is similar to those in type III systems. The inner diameters of the two most likely ring models are large enough to accommodate several copies of other integral membrane proteins. SpoIIQ contains a LytM domain, which is found in metalloendopeptidases, but lacks residues important for metalloprotease activity. Other LytM domains appear to be involved in protein-protein interactions. We found that the LytM domain of SpoIIQ contains an accessory region that interacts with SpoIIIAH.
- Department of Microbiology and Immunology, Emory University School of Medicine, Atlanta, GA 30322, USA.
Organizational Affiliation: