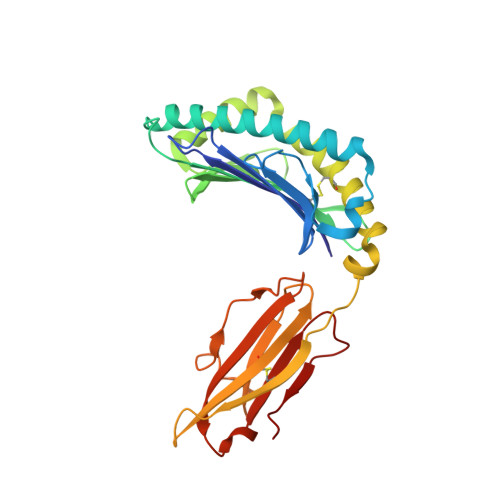
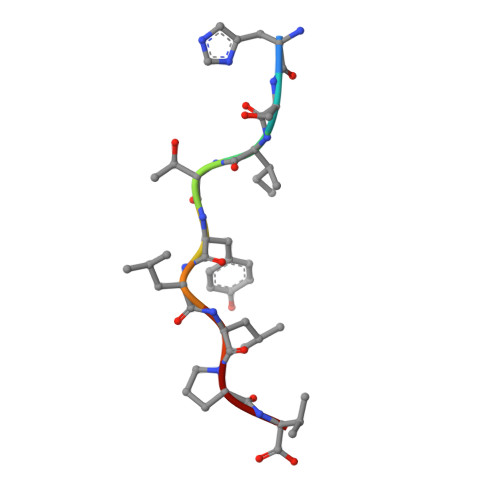
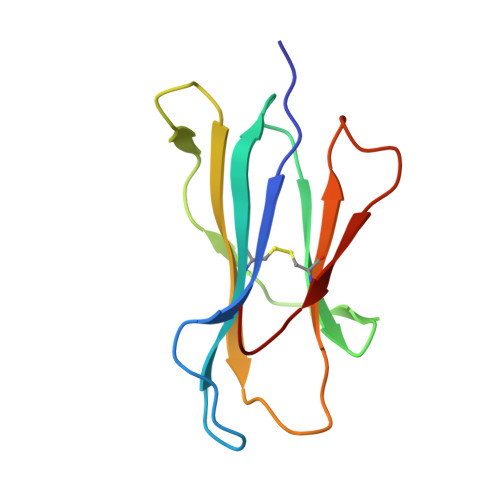
Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire.
Ostrov, D.A., Grant, B.J., Pompeu, Y.A., Sidney, J., Harndahl, M., Southwood, S., Oseroff, C., Lu, S., Jakoncic, J., de Oliveira, C.A., Yang, L., Mei, H., Shi, L., Shabanowitz, J., English, A.M., Wriston, A., Lucas, A., Phillips, E., Mallal, S., Grey, H.M., Sette, A., Hunt, D.F., Buus, S., Peters, B.(2012) Proc Natl Acad Sci U S A 109: 9959-9964
- PubMed: 22645359
- DOI: https://doi.org/10.1073/pnas.1207934109
- Primary Citation of Related Structures:
3UPR - PubMed Abstract:
Idiosyncratic adverse drug reactions are unpredictable, dose-independent and potentially life threatening; this makes them a major factor contributing to the cost and uncertainty of drug development. Clinical data suggest that many such reactions involve immune mechanisms, and genetic association studies have identified strong linkages between drug hypersensitivity reactions to several drugs and specific HLA alleles. One of the strongest such genetic associations found has been for the antiviral drug abacavir, which causes severe adverse reactions exclusively in patients expressing the HLA molecular variant B*57:01. Abacavir adverse reactions were recently shown to be driven by drug-specific activation of cytokine-producing, cytotoxic CD8(+) T cells that required HLA-B*57:01 molecules for their function; however, the mechanism by which abacavir induces this pathologic T-cell response remains unclear. Here we show that abacavir can bind within the F pocket of the peptide-binding groove of HLA-B*57:01, thereby altering its specificity. This provides an explanation for HLA-linked idiosyncratic adverse drug reactions, namely that drugs can alter the repertoire of self-peptides presented to T cells, thus causing the equivalent of an alloreactive T-cell response. Indeed, we identified specific self-peptides that are presented only in the presence of abacavir and that were recognized by T cells of hypersensitive patients. The assays that we have established can be applied to test additional compounds with suspected HLA-linked hypersensitivities in vitro. Where successful, these assays could speed up the discovery and mechanistic understanding of HLA-linked hypersensitivities, and guide the development of safer drugs.
- Department of Pathology, Immunology and Laboratory Medicine, University of Florida College of Medicine, Gainesville, FL 32611, USA.
Organizational Affiliation: