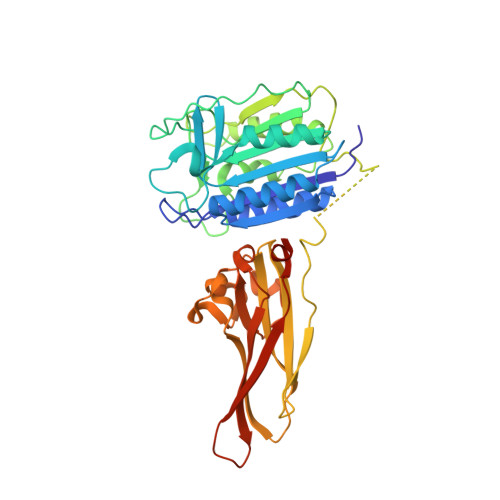
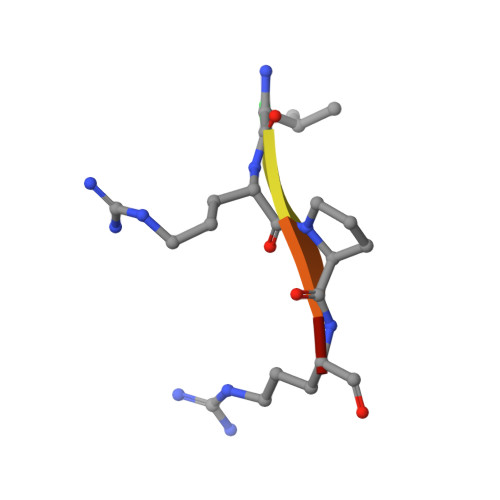
Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Yu, J.W., Jeffrey, P.D., Ha, J.Y., Yang, X., Shi, Y.(2011) Proc Natl Acad Sci U S A 108: 21004-21009
- PubMed: 22158899
- DOI: https://doi.org/10.1073/pnas.1111708108
- Primary Citation of Related Structures:
3UO8, 3UOA - PubMed Abstract:
The mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase, a key component of the Carma1/Bcl10/MALT1 signalosome, is critical for NF-κB signaling in multiple contexts. MALT1 is thought to function as a scaffold and protease to promote signaling; however, the biochemical and structural basis of paracaspase action remains largely unknown. Here we report the 1.75-Å resolution crystal structure of the MALT1 paracaspase region, which contains the paracaspase domain and an ensuing Ig-like domain. The paracaspase and the Ig domains appear as a single folding unit and interact with each other through extensive van der Waals contacts and hydrogen bonds. The paracaspase domain adopts a fold that is nearly identical to that of classic caspases and homodimerizes similarly to form an active protease. Unlike caspases, the active and mature form of the paracaspase domain remains a single uncleaved polypeptide and specifically recognizes the bound peptide inhibitor Val-Arg-Pro-Arg. In particular, the carboxyl-terminal amino acid Arg of the inhibitor is coordinated by three highly conserved acidic residues. This structure serves as an important framework for deciphering the function and mechanism of paracaspases exemplified by MALT1.
- Department of Molecular Biology, Lewis Thomas Laboratory, Princeton University, Princeton, NJ 08544, USA. jong.x.yu@gsk.com
Organizational Affiliation: