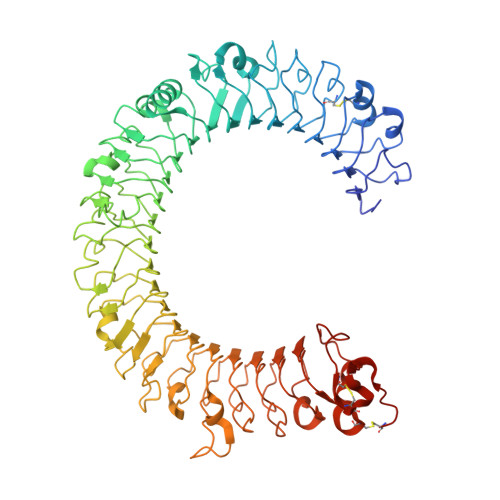
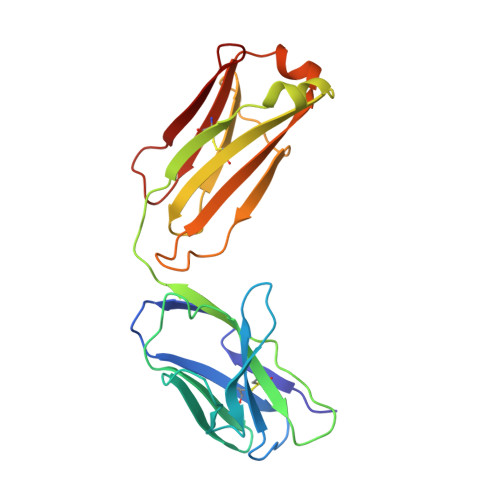
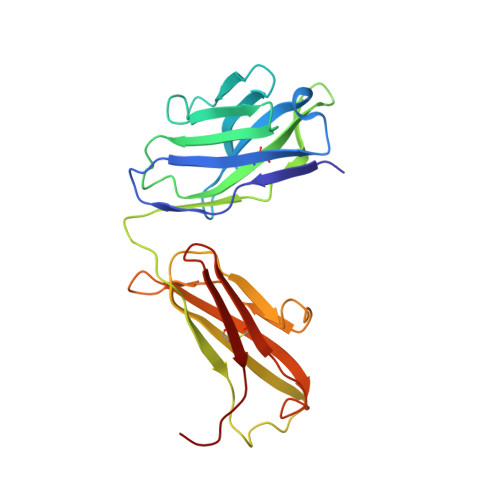
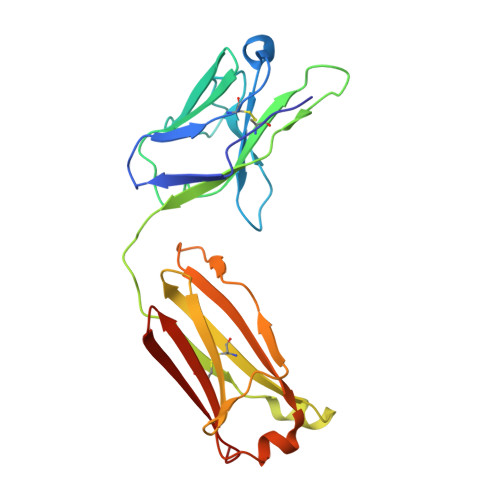
Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
Luo, J., Obmolova, G., Malia, T.J., Wu, S.J., Duffy, K.E., Marion, J.D., Bell, J.K., Ge, P., Zhou, Z.H., Teplyakov, A., Zhao, Y., Lamb, R.J., Jordan, J.L., San Mateo, L.R., Sweet, R.W., Gilliland, G.L.(2012) J Mol Biology 421: 112-124
- PubMed: 22579623
- DOI: https://doi.org/10.1016/j.jmb.2012.05.006
- Primary Citation of Related Structures:
3ULS, 3ULU, 3ULV - PubMed Abstract:
Toll-like receptor 3 (TLR3) recognizes dsRNA and initiates an innate immune response through the formation of a signaling unit (SU) composed of one double-stranded RNA (dsRNA) and two TLR3 molecules. We report the crystal structure of human TLR3 ectodomain (TLR3ecd) in a quaternary complex with three neutralizing Fab fragments. Fab15 binds an epitope that overlaps the C-terminal dsRNA binding site and, in biochemical assays, blocks the interaction of TLR3ecd with dsRNA, thus directly antagonizing TLR3 signaling through inhibition of SU formation. In contrast, Fab12 and Fab1068 bind TLR3ecd at sites distinct from the N- and C-terminal regions that interact with dsRNA and do not inhibit minimal SU formation with short dsRNA. Molecular modeling based on the co-structure rationalizes these observations by showing that both Fab12 and Fab1068 prevent lateral clustering of SUs along the length of the dsRNA ligand. This model is further supported by cell-based assay results using dsRNA ligands of lengths that support single and multiple SUs. Thus, their antagonism of TLR3 signaling indicates that lateral clustering of SUs is required for TLR3 signal transduction.
- Biologics Research, Janssen Research and Development, L.L.C., 145 King of Prussia Road, Radnor, PA 19087, USA. jluo@its.jnj.com
Organizational Affiliation: