Structure of Bos taurus Arp2/3 complex with bound inhibitor CK-869 and ATP
Nolen, B.J., Han, M.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 3 | 418 | Bos taurus | Mutation(s): 0 | 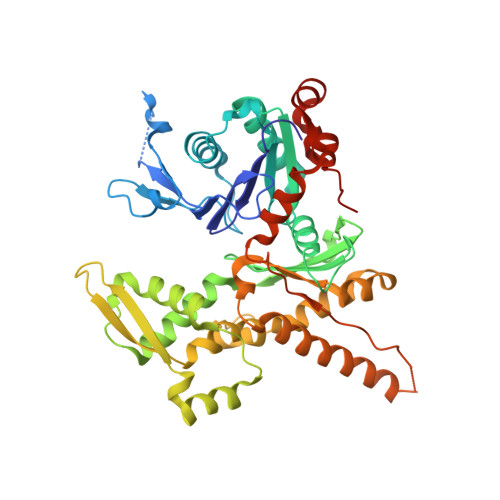 | |
UniProt | |||||
Find proteins for P61157 (Bos taurus) Explore P61157 Go to UniProtKB: P61157 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61157 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2 | 394 | Bos taurus | Mutation(s): 0 | 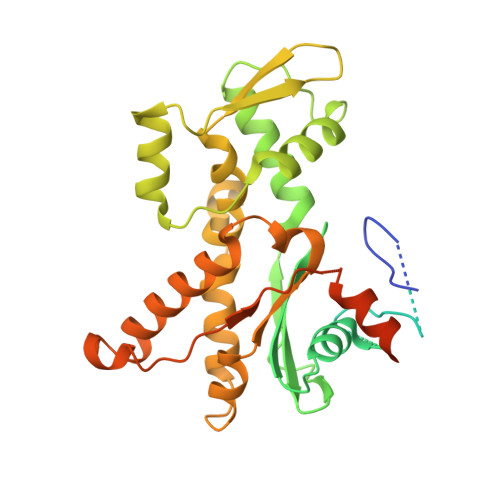 | |
UniProt | |||||
Find proteins for A7MB62 (Bos taurus) Explore A7MB62 Go to UniProtKB: A7MB62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A7MB62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2/3 complex subunit 1B | 372 | Bos taurus | Mutation(s): 0 | 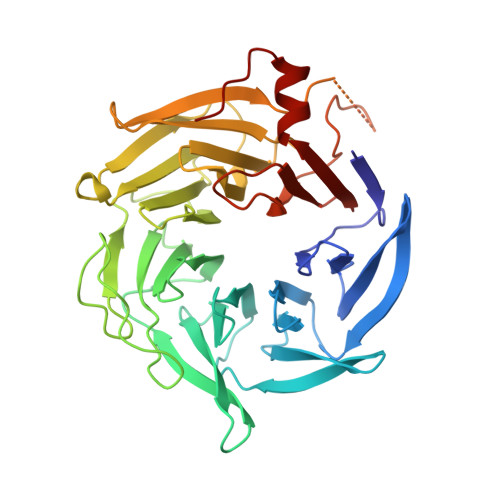 | |
UniProt | |||||
Find proteins for Q58CQ2 (Bos taurus) Explore Q58CQ2 Go to UniProtKB: Q58CQ2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q58CQ2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2/3 complex subunit 2 | 300 | Bos taurus | Mutation(s): 0 | 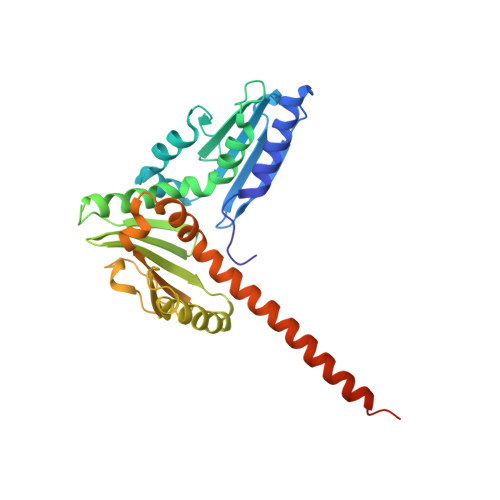 | |
UniProt | |||||
Find proteins for Q3MHR7 (Bos taurus) Explore Q3MHR7 Go to UniProtKB: Q3MHR7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3MHR7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2/3 complex subunit 3 | 178 | Bos taurus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q3T035 (Bos taurus) Explore Q3T035 Go to UniProtKB: Q3T035 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3T035 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2/3 complex subunit 4 | 168 | Bos taurus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q148J6 (Bos taurus) Explore Q148J6 Go to UniProtKB: Q148J6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q148J6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actin-related protein 2/3 complex subunit 5 | 151 | Bos taurus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for Q3SYX9 (Bos taurus) Explore Q3SYX9 Go to UniProtKB: Q3SYX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3SYX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | H [auth A], K [auth B] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| C69 Query on C69 | I [auth A] | (2S)-2-(3-bromophenyl)-3-(2,4-dimethoxyphenyl)-1,3-thiazolidin-4-one C17 H16 Br N O3 S MVWNPZYLNLATCH-KRWDZBQOSA-N |  | ||
| CA Query on CA | J [auth A], L [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 111.166 | α = 90 |
| b = 128.314 | β = 90 |
| c = 198.492 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| CNS | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| CNS | phasing |