Structural basis of the drug-binding specificity of human serum albumin
Curry, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serum albumin | A, B [auth H] | 585 | Homo sapiens | Mutation(s): 0 | 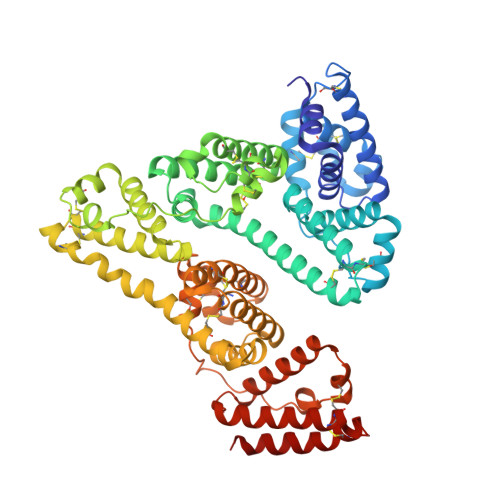 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P02768 (Homo sapiens) Explore P02768 Go to UniProtKB: P02768 | |||||
PHAROS: P02768 GTEx: ENSG00000163631 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MYR Query on MYR | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | MYRISTIC ACID C14 H28 O2 TUNFSRHWOTWDNC-UHFFFAOYSA-N |  | ||
| 308 Query on 308 | P [auth H] | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine C10 H17 N DKNWSYNQZKUICI-CHIWXEEVSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 38.27 | α = 74.91 |
| b = 93.516 | β = 89.75 |
| c = 96.217 | γ = 80.04 |
| Software Name | Purpose |
|---|---|
| CBASS | data collection |
| AMoRE | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | refinement |