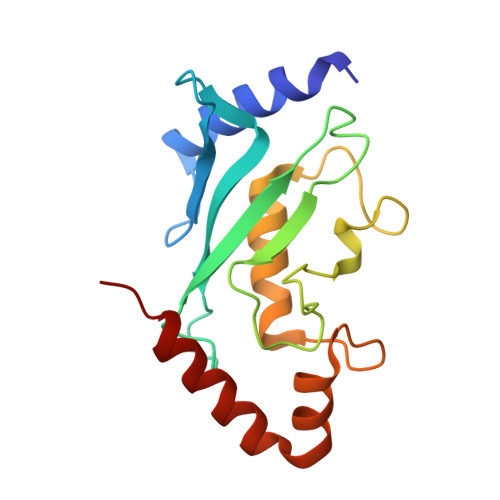
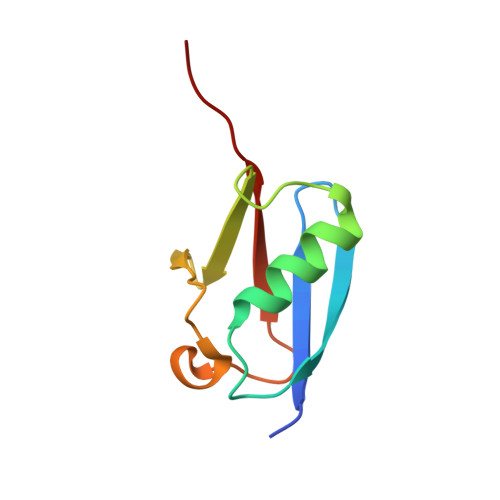
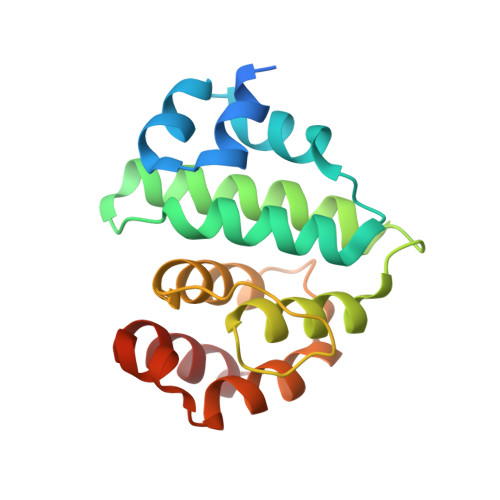
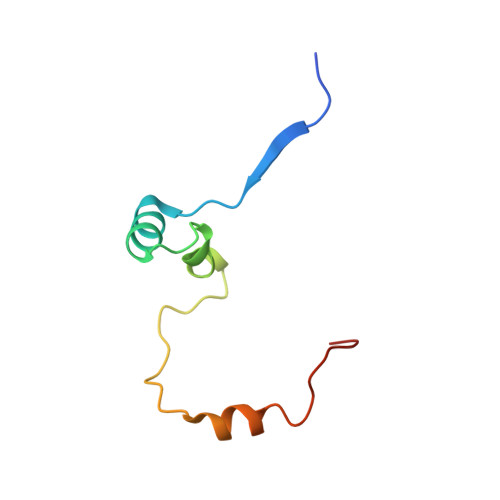
Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
Gareau, J.R., Reverter, D., Lima, C.D.(2012) J Biological Chem 287: 4740-4751
- PubMed: 22194619
- DOI: https://doi.org/10.1074/jbc.M111.321141
- Primary Citation of Related Structures:
3UIN, 3UIO, 3UIP - PubMed Abstract:
The RanBP2 nucleoporin contains an internal repeat domain (IR1-M-IR2) that catalyzes E3 ligase activity and forms a stable complex with SUMO-modified RanGAP1 and UBC9 at the nuclear pore complex. RanBP2 exhibits specificity for SUMO1 as RanGAP1-SUMO1/UBC9 forms a more stable complex with RanBP2 compared with RanGAP1-SUMO2 that results in greater protection of RanGAP-SUMO1 from proteases. The IR1-M-IR2 SUMO E3 ligase activity also shows a similar preference for SUMO1. We utilized deletions and domain swap constructs in protease protection assays and automodification assays to define RanBP2 domains responsible for RanGAP1-SUMO1 protection and SUMO1-specific E3 ligase activity. Our data suggest that elements in both IR1 and IR2 exhibit specificity for SUMO1. IR1 protects RanGAP1-SUMO1/UBC9 and functions as the primary E3 ligase of RanBP2, whereas IR2 retains the ability to interact with SUMO1 to promote SUMO1-specific E3 ligase activity. To determine the structural basis for SUMO1 specificity, a hybrid IR1 construct and IR1 were used to determine three new structures for complexes containing UBC9 with RanGAP1-SUMO1/2. These structures show more extensive contacts among SUMO, UBC9, and RanBP2 in complexes containing SUMO1 compared with SUMO2 and suggest that differences in SUMO specificity may be achieved through these subtle conformational differences.
- Structural Biology Program, Sloan-Kettering Institute, New York, New York 10065, USA.
Organizational Affiliation: