Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent.
Andraos, R., Qian, Z., Bonenfant, D., Rubert, J., Vangrevelinghe, E., Scheufler, C., Marque, F., Regnier, C.H., De Pover, A., Ryckelynck, H., Bhagwat, N., Koppikar, P., Goel, A., Wyder, L., Tavares, G., Baffert, F., Pissot-Soldermann, C., Manley, P.W., Gaul, C., Voshol, H., Levine, R.L., Sellers, W.R., Hofmann, F., Radimerski, T.(2012) Cancer Discov 2: 512-523
- PubMed: 22684457
- DOI: https://doi.org/10.1158/2159-8290.CD-11-0324
- Primary Citation of Related Structures:
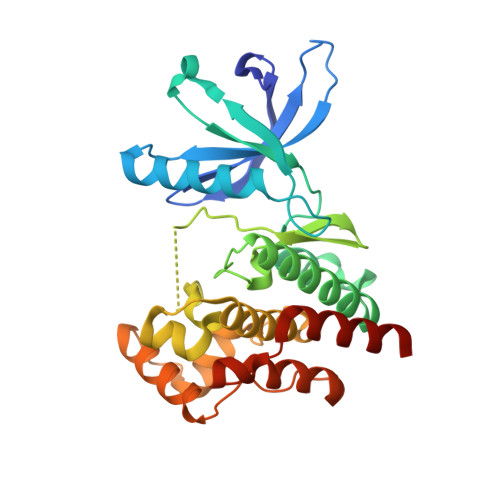
3UGC - PubMed Abstract:
Janus kinase (JAK) inhibitors are being developed for the treatment of rheumatoid arthritis, psoriasis, myeloproliferative neoplasms, and leukemias. Most of these drugs target the ATP-binding pocket and stabilize the active conformation of the JAK kinases. This type I binding mode can lead to an increase in JAK activation loop phosphorylation, despite blockade of kinase function. Here we report that stabilizing the inactive state via type II inhibition acts in the opposite manner, leading to a loss of activation loop phosphorylation. We used X-ray crystallography to corroborate the binding mode and report for the first time the crystal structure of the JAK2 kinase domain in an inactive conformation. Importantly, JAK inhibitor-induced activation loop phosphorylation requires receptor interaction, as well as intact kinase and pseudokinase domains. Hence, depending on the respective conformation stabilized by a JAK inhibitor, hyperphosphorylation of the activation loop may or may not be elicited.
- Disease Area Oncology, Novartis Institutes for BioMedical Research, Basel, Switzerland.
Organizational Affiliation: