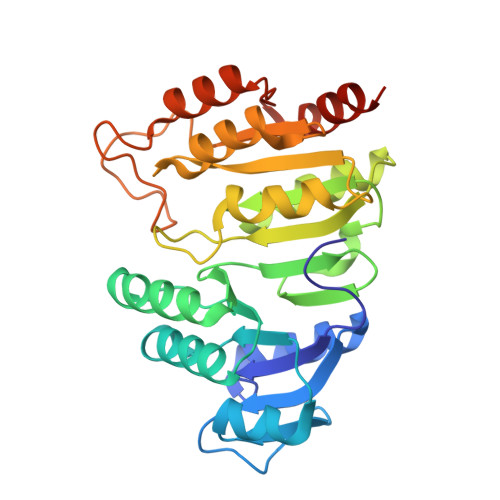
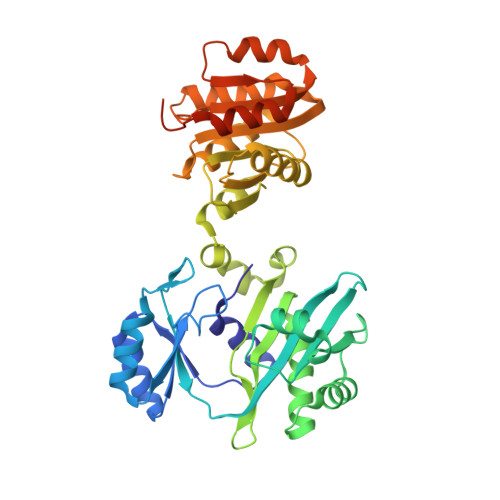
Biochemical and structural characterization of the GTP-preferring succinyl-CoA synthetase from Thermus aquaticus.
Joyce, M.A., Hayakawa, K., Wolodko, W.T., Fraser, M.E.(2012) Acta Crystallogr D Biol Crystallogr 68: 751-762
- PubMed: 22751660
- DOI: https://doi.org/10.1107/S0907444912010852
- Primary Citation of Related Structures:
3UFX - PubMed Abstract:
Succinyl-CoA synthetase (SCS) from Thermus aquaticus was characterized biochemically via measurements of the activity of the enzyme and determination of its quaternary structure as well as its stability and refolding properties. The enzyme is most active between pH 8.0 and 8.4 and its activity increases with temperature to about 339 K. Gel-filtration chromatography and sedimentation equilibrium under native conditions demonstrated that the enzyme is a heterotetramer of two α-subunits and two β-subunits. The activity assays showed that the enzyme uses either ADP/ATP or GDP/GTP, but prefers GDP/GTP. This contrasts with Escherichia coli SCS, which uses GDP/GTP but prefers ADP/ATP. To understand the nucleotide preference, T. aquaticus SCS was crystallized in the presence of GDP, leading to the determination of the structure in complex with GDP-Mn(2+). A water molecule and Pro20β in T. aquaticus take the place of Gln20β in pig GTP-specific SCS, interacting well with the guanine base and other residues of the nucleotide-binding site. This leads to the preference for GDP/GTP, but does not hinder the binding of ADP/ATP.
- Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.
Organizational Affiliation: