Structure of the complex between CbpA J-domain and CbpM provides a link between chaperone and transcription regulation in bacterial heat shock response
Sarraf, N.S., Shi, R., Zhang, L., Cygler, M., Ekiel, I.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chaperone-modulator protein CbpM | 102 | Klebsiella variicola 342 | Mutation(s): 0 Gene Names: cbpM, KPK_5158 | 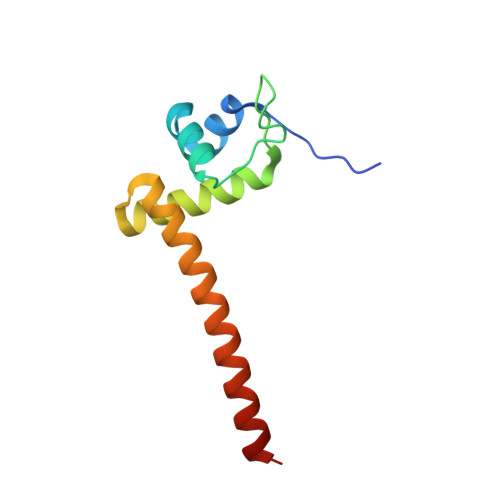 | |
UniProt | |||||
Find proteins for B5Y388 (Klebsiella variicola (strain 342)) Explore B5Y388 Go to UniProtKB: B5Y388 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B5Y388 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Curved DNA-binding protein | 74 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: b1000, cbpA, JW0985 | 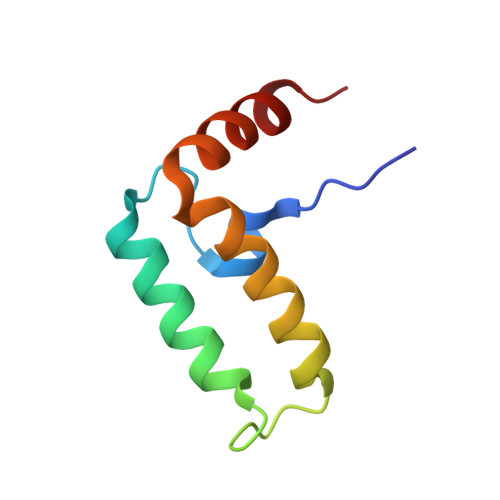 | |
UniProt | |||||
Find proteins for P36659 (Escherichia coli (strain K12)) Explore P36659 Go to UniProtKB: P36659 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P36659 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 52.816 | α = 90 |
| b = 77.094 | β = 90 |
| c = 111.231 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| SHARP | phasing |