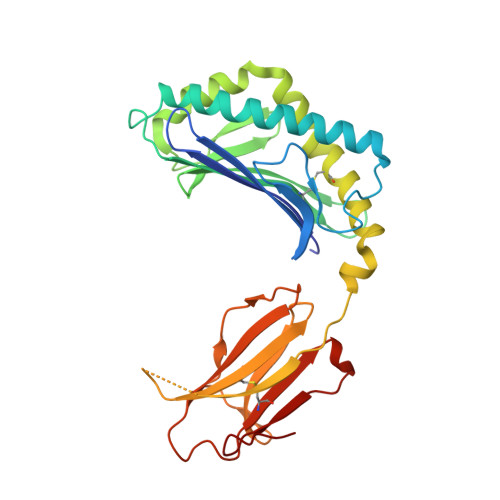
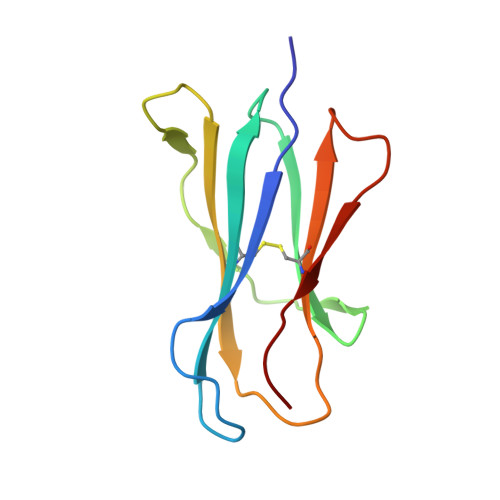
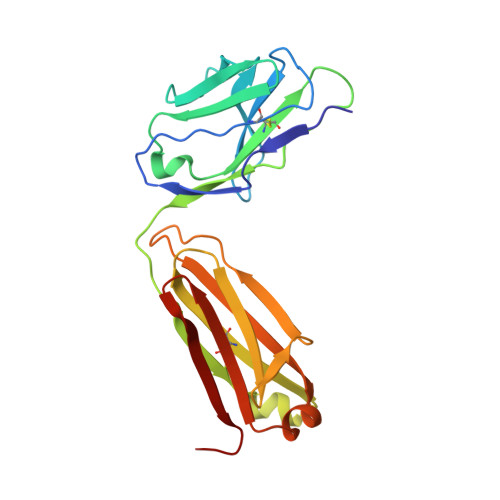
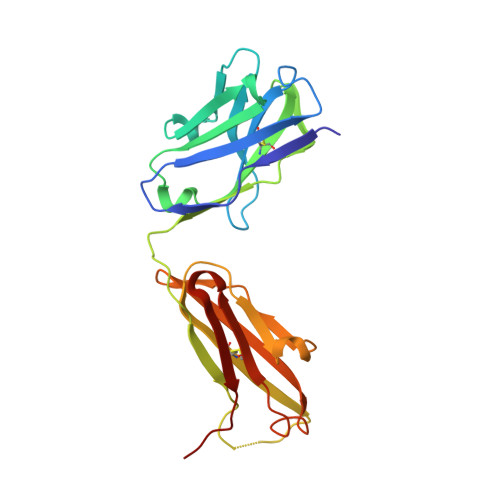
Structural basis for the recognition of C20:2-alpha GalCer by the invariant natural killer T cell receptor-like antibody L363.
Yu, E.D., Girardi, E., Wang, J., Mac, T.T., Yu, K.O., Van Calenbergh, S., Porcelli, S.A., Zajonc, D.M.(2012) J Biological Chem 287: 1269-1278
- PubMed: 22110136
- DOI: https://doi.org/10.1074/jbc.M111.308783
- Primary Citation of Related Structures:
3UBX - PubMed Abstract:
Natural killer T (NKT) cells express a semi-invariant Vα14 T cell receptor (TCR) and recognize structurally diverse antigens presented by the antigen-presenting molecule CD1d that range from phosphoglycerolipids to α- and β-anomeric glycosphingolipids, as well as microbial α-glycosyl diacylglycerolipids. Recently developed antibodies that are specific for the complex of the prototypical invariant NKT (iNKT) cell antigen αGalCer (KRN7000) bound to mouse CD1d have become valuable tools in elucidating the mechanism of antigen loading and presentation. Here, we report the 3.1 Å resolution crystal structure of the Fab of one of these antibodies, L363, bound to mCD1d complexed with the αGalCer analog C20:2, revealing that L363 is an iNKT TCR-like antibody that binds CD1d-presented αGalCer in a manner similar to the TCR. The structure reveals that L363 depends on both the L and H chains for binding to the glycolipid-mCD1d complex, although only the L chain is involved in contacts with the glycolipid antigen. The H chain of L363 features residue Trp-104, which mimics the TCR CDR3α residue Leu-99, which is crucial for CD1d binding. We characterized the antigen-specificity of L363 toward several different glycolipids, demonstrating that whereas the TCR can induce structural changes in both antigen and CD1d to recognize disparate lipid antigens, the antibody L363 can only induce the F' roof formation in CD1d but fails to reorient the glycolipid headgroup necessary for binding. In summary, L363 is a powerful tool to study mechanism of iNKT cell activation for structural analogs of KRN7000, and our study can aid in the design of antibodies with altered antigen specificity.
- Division of Cell Biology, La Jolla Institute for Allergy & Immunology, La Jolla, California 92037, USA.
Organizational Affiliation: