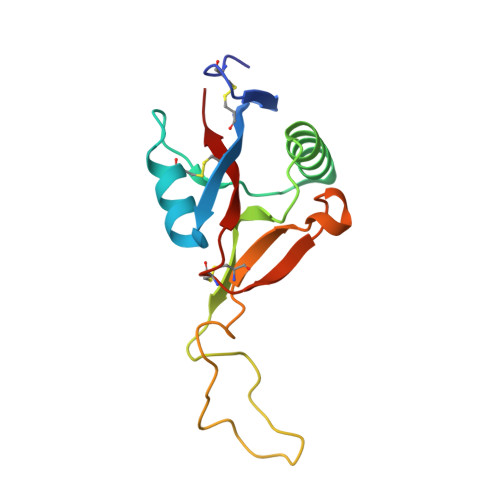
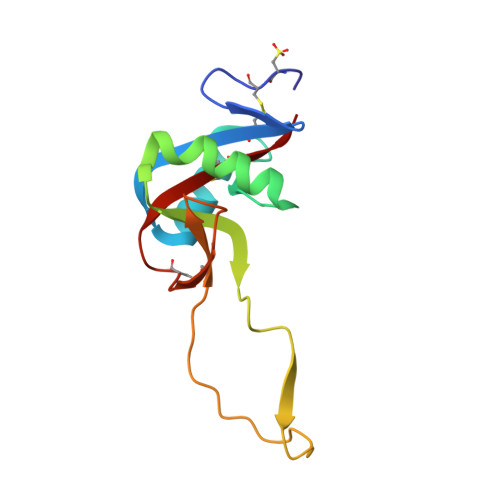
Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation.
Gao, Y., Ge, H., Chen, H., Li, H., Liu, Y., Chen, L., Li, X., Liu, J., Niu, L., Teng, M.(2012) Proteins
- PubMed: 22447656
- DOI: https://doi.org/10.1002/prot.24060
- Primary Citation of Related Structures:
3UBU - PubMed Abstract:
Agkisacucetin is a snake C-type lectin isolated from the venom of Agkistrodon acutus (A. acutus). It binds specifically to the platelet glycoprotein (GP) Ib and prevents the von Willebrand factor (VWF) accessing it. We determined the crystal structure of agkisacucetin to 1.9Å resolution. The structure of agkisacucetin has an (αβ) fold similar to another GPIb-binding protein, flavocetin-A, but lacks the C-terminal cysteine in the β-subunit, does not form (βα)(4) tetramers, and does not cluster GPIbs, like flavocetin-A.
- Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: