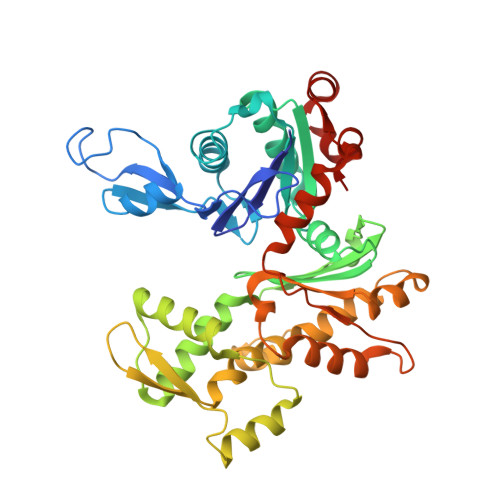
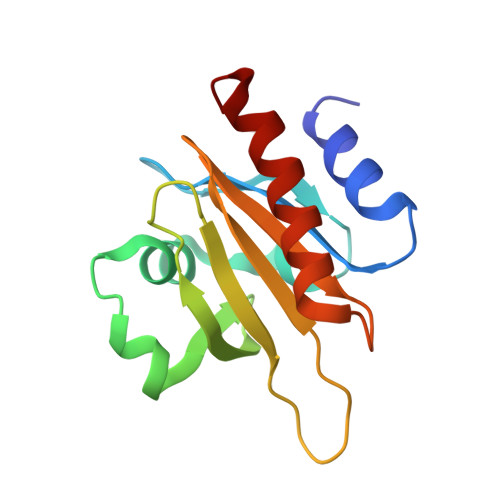
Structural basis for profilin-mediated actin nucleotide exchange.
Porta, J.C., Borgstahl, G.E.(2012) J Mol Biology 418: 103-116
- PubMed: 22366544
- DOI: https://doi.org/10.1016/j.jmb.2012.02.012
- Primary Citation of Related Structures:
3U4L, 3UB5 - PubMed Abstract:
Actin is a ubiquitous eukaryotic protein that is responsible for cellular scaffolding, motility, and division. The ability of actin to form a helical filament is the driving force behind these cellular activities. Formation of a filament depends on the successful exchange of actin's ADP for ATP. Mammalian profilin is a small actin binding protein that catalyzes the exchange of nucleotide and facilitates the addition of an actin monomer to a growing filament. Here, crystal structures of profilin-actin have been determined to show an actively exchanging ATP. Structural analysis shows how the binding of profilin to the barbed end of actin causes a rotation of the small domain relative to the large domain. This conformational change is propagated to the ATP site and causes a shift in nucleotide loops, which in turn causes a repositioning of Ca(2+) to its canonical position as the cleft closes around ATP. Reversal of the solvent exposure of Trp356 is also involved in cleft closure. In addition, secondary calcium binding sites were identified.
- Department of Biochemistry and Molecular Biology, 987696 Nebraska Medical Center, Omaha, NE 68198-7696, USA.
Organizational Affiliation: