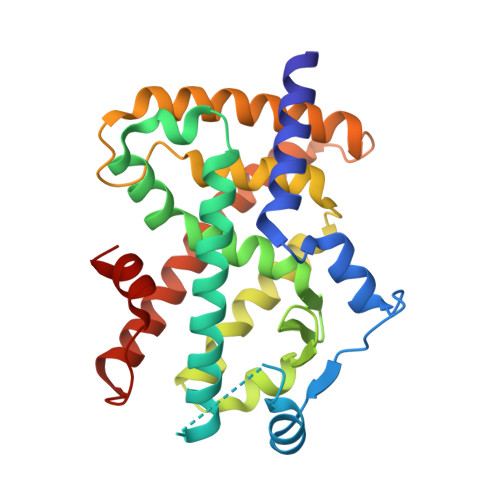
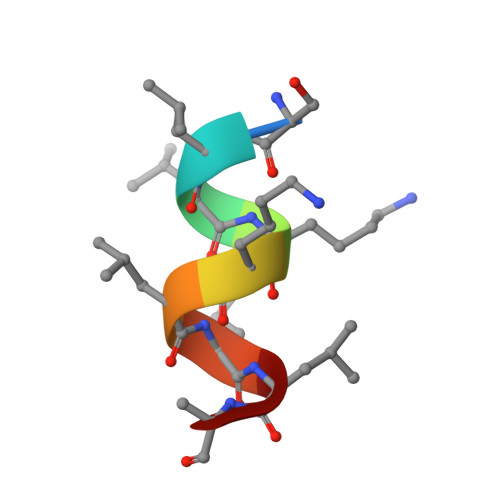
Identification and Mechanism of 10-Carbon Fatty Acid as Modulating Ligand of Peroxisome Proliferator-activated Receptors.
Malapaka, R.R., Khoo, S., Zhang, J., Choi, J.H., Zhou, X.E., Xu, Y., Gong, Y., Li, J., Yong, E.L., Chalmers, M.J., Chang, L., Resau, J.H., Griffin, P.R., Chen, Y.E., Xu, H.E.(2012) J Biological Chem 287: 183-195
- PubMed: 22039047
- DOI: https://doi.org/10.1074/jbc.M111.294785
- Primary Citation of Related Structures:
3U9Q - PubMed Abstract:
Peroxisome proliferator-activated receptors (PPARα, -β/δ, and -γ) are a subfamily of nuclear receptors that plays key roles in glucose and lipid metabolism. PPARγ is the molecular target of the thiazolidinedione class of antidiabetic drugs that has many side effects. PPARγ is also activated by long chain unsaturated or oxidized/nitrated fatty acids, but its relationship with the medium chain fatty acids remains unclear even though the medium chain triglyceride oils have been used to control weight gain and glycemic index. Here, we show that decanoic acid (DA), a 10-carbon fatty acid and a major component of medium chain triglyceride oils, is a direct ligand of PPARγ. DA binds and partially activates PPARγ without leading to adipogenesis. Crystal structure reveals that DA occupies a novel binding site and only partially stabilizes the AF-2 helix. DA also binds weakly to PPARα and PPARβ/δ. Treatments with DA and its triglyceride form improve glucose sensitivity and lipid profiles without weight gain in diabetic mice. Together, these results suggest that DA is a modulating ligand for PPARs, and the structure can aid in designing better and safer PPARγ-based drugs.
- Laboratories of Structural Sciences, Van Andel Research Institute, Grand Rapids, Michigan, 49503. Electronic address: vmalapaka@rics.bwh.harvard.edu.
Organizational Affiliation: