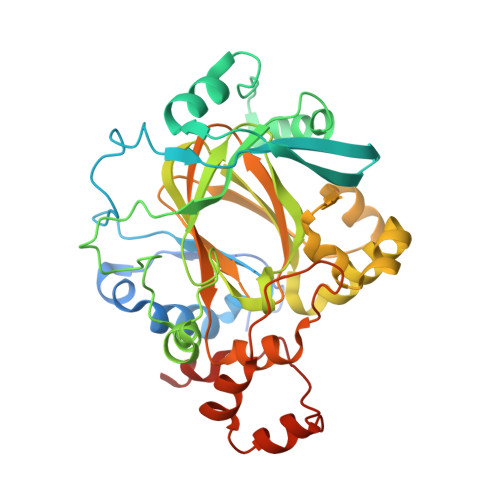
Linking of 2-Oxoglutarate and Substrate Binding Sites Enables Potent and Highly Selective Inhibition of JmjC Histone Demethylases.
Woon, E.C., Tumber, A., Kawamura, A., Hillringhaus, L., Ge, W., Rose, N.R., Ma, J.H., Chan, M.C., Walport, L.J., Che, K.H., Ng, S.S., Marsden, B.D., Oppermann, U., McDonough, M.A., Schofield, C.J.(2012) Angew Chem Int Ed Engl 51: 1631-1634
- PubMed: 22241642
- DOI: https://doi.org/10.1002/anie.201107833
- Primary Citation of Related Structures:
3U4S - Chemistry Research Laboratory, University of Oxford, 12 Mansfield Road, Oxford, OX1 3TA, UK.
Organizational Affiliation: