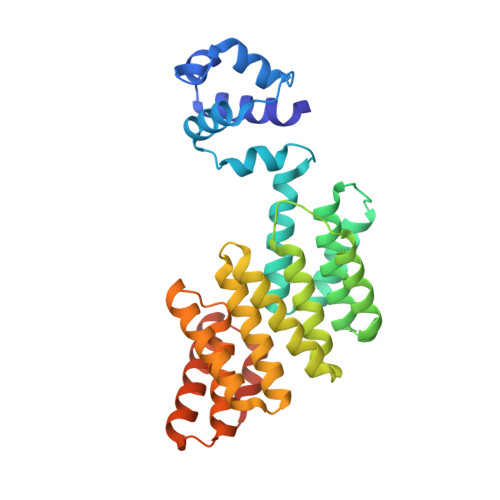
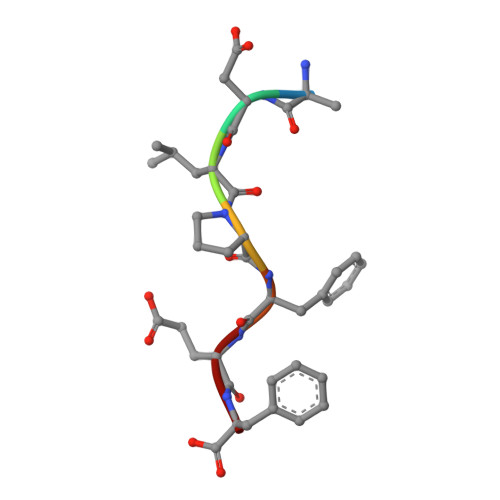
Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR.
Grenha, R., Slamti, L., Nicaise, M., Refes, Y., Lereclus, D., Nessler, S.(2013) Proc Natl Acad Sci U S A 110: 1047-1052
- PubMed: 23277548
- DOI: https://doi.org/10.1073/pnas.1213770110
- Primary Citation of Related Structures:
3U3W, 4FSC - PubMed Abstract:
The quorum-sensing regulator PlcR is the master regulator of most known virulence factors in Bacillus cereus. It is a helix-turn-helix (HTH)-type transcription factor activated upon binding of its cognate signaling peptide PapR on a tetratricopeptide repeat-type regulatory domain. The structural and functional properties of PlcR have defined a new family of sensor regulators, called the RNPP family (for Rap, NprR, PrgX, and PlcR), in Gram-positive bacteria. To fully understand the activation mechanism of PlcR, we took a closer look at the conformation changes induced upon binding of PapR and of its target DNA, known as PlcR-box. For that purpose we have determined the structures of the apoform of PlcR (Apo PlcR) and of the ternary complex of PlcR with PapR and the PlcR-box from the plcA promoter. Comparison of the apoform of PlcR with the previously published structure of the PlcR-PapR binary complex shows how a small conformational change induced in the C-terminal region of the tetratricopeptide repeat (TPR) domain upon peptide binding propagates via the linker helix to the N-terminal HTH DNA-binding domain. Further comparison with the PlcR-PapR-DNA ternary complex shows how the activation of the PlcR dimer allows the linker helix to undergo a drastic conformational change and subsequent proper positioning of the HTH domains in the major groove of the two half sites of the pseudopalindromic PlcR-box. Together with random mutagenesis experiments and interaction measurements using peptides from distinct pherogroups, this structural analysis allows us to propose a molecular mechanism for this functional switch.
- Laboratoire d'Enzymologie et Biochimie Structurales, Unité Propre de Recherche 3082, Centre National de la Recherche Scientifique, 91198 Gif sur Yvette, France.
Organizational Affiliation: