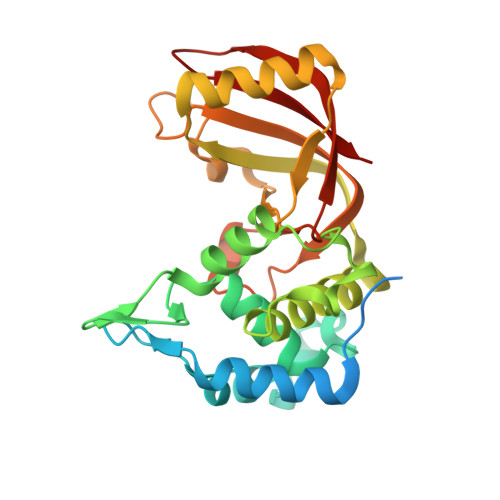
Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity
Jeong, B.-R., Lin, Y., Joe, A., Guo, M., Korneli, C., Yang, H., Wang, P., Yu, M., Cerny, R.L., Staiger, D., Alfano, J.R., Xu, Y.(2011) J Biological Chem 286: 43272-43281
- PubMed: 22013065
- DOI: https://doi.org/10.1074/jbc.M111.290122
- Primary Citation of Related Structures:
3U0J - PubMed Abstract:
The Pseudomonas syringae type III effector HopU1 is a mono-ADP-ribosyltransferase that is injected into plant cells by the type III protein secretion system. Inside the plant cell it suppresses immunity by modifying RNA-binding proteins including the glycine-rich RNA-binding protein GRP7. The crystal structure of HopU1 at 2.7-Å resolution reveals two unique protruding loops, L1 and L4, not found in other mono-ADP-ribosyltransferases. Site-directed mutagenesis demonstrates that these loops are essential for substrate recognition and enzymatic activity. HopU1 ADP-ribosylates the conserved arginine 49 of GRP7, and this reduces the ability of GRP7 to bind RNA in vitro. In vivo, expression of GRP7 with Arg-49 replaced with lysine does not complement the reduced immune responses of the Arabidopsis thaliana grp7-1 mutant demonstrating the importance of this residue for GRP7 function. These data provide mechanistic details how HopU1 recognizes this novel type of substrate and highlights the role of GRP7 in plant immunity.
- Center for Plant Science Innovation and the Department of Plant Pathology, University of Nebraska, Lincoln, Nebraska 68588-0660, USA.
Organizational Affiliation: