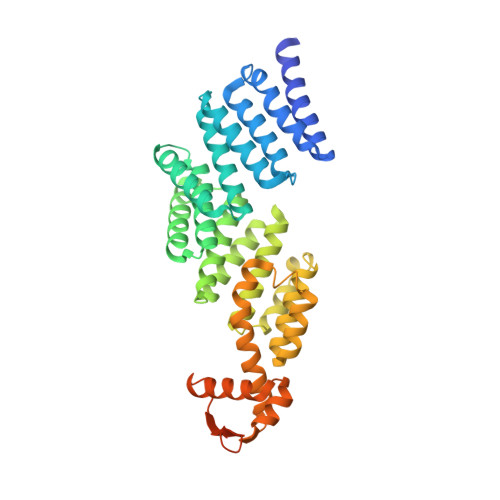
The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together.
Pathare, G.R., Nagy, I., Bohn, S., Unverdorben, P., Hubert, A., Korner, R., Nickell, S., Lasker, K., Sali, A., Tamura, T., Nishioka, T., Forster, F., Baumeister, W., Bracher, A.(2012) Proc Natl Acad Sci U S A 109: 149-154
- PubMed: 22187461
- DOI: https://doi.org/10.1073/pnas.1117648108
- Primary Citation of Related Structures:
3TXM, 3TXN - PubMed Abstract:
Proteasomes execute the degradation of most cellular proteins. Although the 20S core particle (CP) has been studied in great detail, the structure of the 19S regulatory particle (RP), which prepares ubiquitylated substrates for degradation, has remained elusive. Here, we report the crystal structure of one of the RP subunits, Rpn6, and we describe its integration into the cryo-EM density map of the 26S holocomplex at 9.1 Å resolution. Rpn6 consists of an α-solenoid-like fold and a proteasome COP9/signalosome eIF3 (PCI) module in a right-handed suprahelical configuration. Highly conserved surface areas of Rpn6 interact with the conserved surfaces of the Pre8 (alpha2) and Rpt6 subunits from the alpha and ATPase rings, respectively. The structure suggests that Rpn6 has a pivotal role in stabilizing the otherwise weak interaction between the CP and the RP.
- Department of Molecular Structural Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: