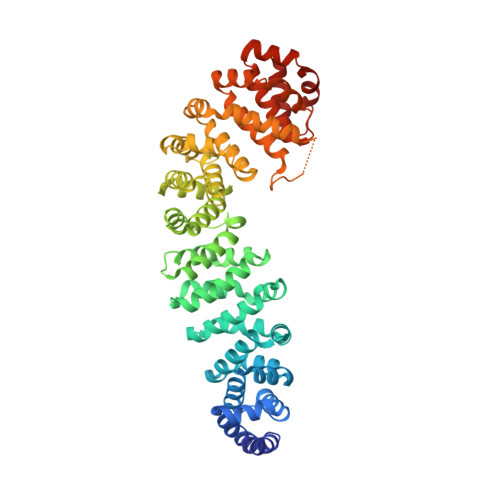
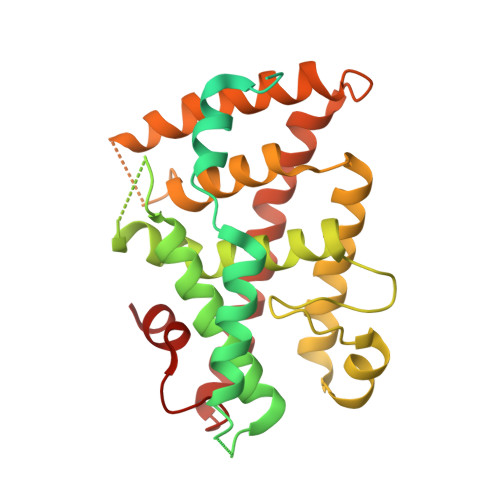
Structural basis of coactivation of liver receptor homolog-1 by beta-catenin.
Yumoto, F., Nguyen, P., Sablin, E.P., Baxter, J.D., Webb, P., Fletterick, R.J.(2012) Proc Natl Acad Sci U S A 109: 143-148
- PubMed: 22187462
- DOI: https://doi.org/10.1073/pnas.1117036108
- Primary Citation of Related Structures:
3TX7 - PubMed Abstract:
We report the three-dimensional structure of a β-catenin armadillo repeat in complex with the liver receptor homolog-1 (LRH-1) ligand binding domain at 2.8 Å resolution as the first structure of β-catenin in complex with any nuclear receptor. The surface of β-catenin that binds LRH-1 partly overlaps defined contact sites for peptide segments of β-catenin partners, including T-cell factor-4. The surface of LRH-1 that engages β-catenin is comprised of helices 1, 9, and 10 and is distinct from known interaction surfaces of LRH-1, including corepressor and coactivator binding sites. Targeted mutagenesis of amino acids forming both sides of the LRH-1/β-catenin interface reveals that they are essential for stable interactions between these proteins in solution. The LRH-1 binding site in β-catenin is also required for association with androgen receptor, providing evidence that the observed LRH-1/β-catenin interaction may be prototypic.
- Department of Biochemistry and Biophysics, University of California San Francisco, 600 16th Street, San Francisco, CA 94158, USA.
Organizational Affiliation: