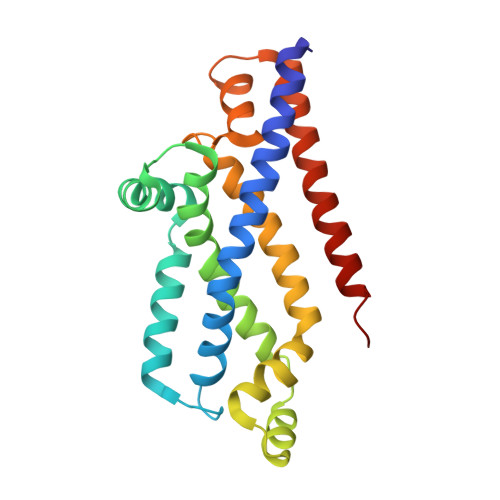
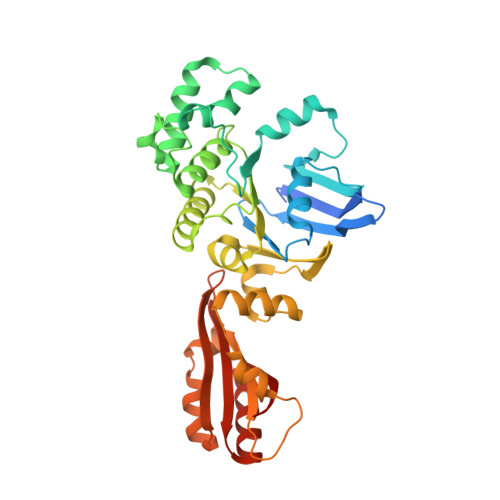
Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition.
Johnson, E., Nguyen, P.T., Yeates, T.O., Rees, D.C.(2012) Protein Sci 21: 84-96
- PubMed: 22095702
- DOI: https://doi.org/10.1002/pro.765
- Primary Citation of Related Structures:
3TUI, 3TUJ, 3TUZ - PubMed Abstract:
Two new crystal structures of the Escherichia coli high affinity methionine uptake ATP Binding Cassette (ABC) transporter MetNI, purified in the detergents cyclohexyl-pentyl-β-D-maltoside (CY5) and n-decyl-β-D-maltopyranoside (DM), have been solved in inward facing conformations to resolutions of 2.9 and 4.0 Å, respectively. Compared to the previously reported 3.7 Å resolution structure of MetNI purified in n-dodecyl-β-D-maltopyranoside (DDM), the higher resolution of the CY5 data enabled significant improvements to the structural model in several regions, including corrections to the sequence registry, and identification of ADP in the nucleotide binding site. CY5 crystals soaked with selenomethionine established details of the methionine binding site in the C2 regulatory domain of the ABC subunit, including the displacement of the side chain of MetN residue methionine 301 by the exogenous ligand. When compared to the CY5 or DDM structures, the DM structure exhibits a significant repositioning of the dimeric C2 domains, including an unexpected register shift in the intermolecular β-sheet hydrogen bonding between monomers, and a narrowing of the nucleotide binding space. The immediate proximity of the exogenous methionine binding site to the conformationally variable dimeric interface provides an indication of how methionine binding to the regulatory domains might mediate the phenomenon of transinhibition.
- Division of Chemistry and Chemical Engineering 114-96, California Institute of Technology, Pasadena, CA 91125, USA.
Organizational Affiliation: