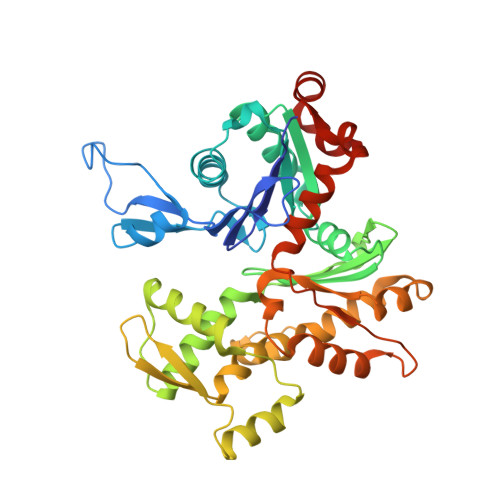
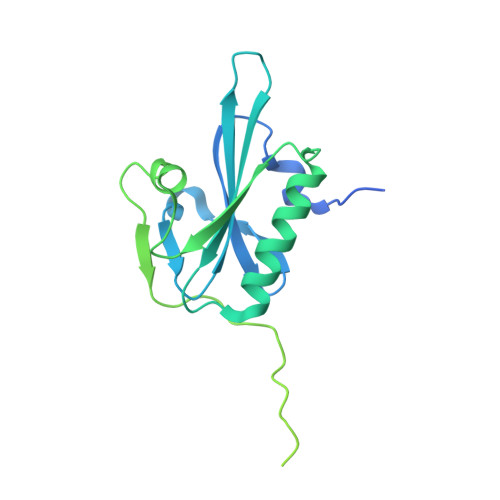
Structural States and dynamics of the d-loop in actin.
Durer, Z.A., Kudryashov, D.S., Sawaya, M.R., Altenbach, C., Hubbell, W., Reisler, E.(2012) Biophys J 103: 930-939
- PubMed: 23009842
- DOI: https://doi.org/10.1016/j.bpj.2012.07.030
- Primary Citation of Related Structures:
3TU5 - PubMed Abstract:
Conformational changes induced by ATP hydrolysis on actin are involved in the regulation of complex actin networks. Previous structural and biochemical data implicate the DNase I binding loop (D-loop) of actin in such nucleotide-dependent changes. Here, we investigated the structural and conformational states of the D-loop (in solution) using cysteine scanning mutagenesis and site-directed labeling. The reactivity of D-loop cysteine mutants toward acrylodan and the mobility of spin labels on these mutants do not show patterns of an α-helical structure in monomeric and filamentous actin, irrespective of the bound nucleotide. Upon transition from monomeric to filamentous actin, acrylodan emission spectra and electron paramagnetic resonance line shapes of labeled mutants are blue-shifted and more immobilized, respectively, with the central residues (residues 43-47) showing the most drastic changes. Moreover, complex electron paramagnetic resonance line shapes of spin-labeled mutants suggest several conformational states of the D-loop. Together with a new (to our knowledge) actin crystal structure that reveals the D-loop in a unique hairpin conformation, our data suggest that the D-loop equilibrates in F-actin among different conformational states irrespective of the nucleotide state of actin.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, California, USA. zeynepdurer@ucla.edu
Organizational Affiliation: