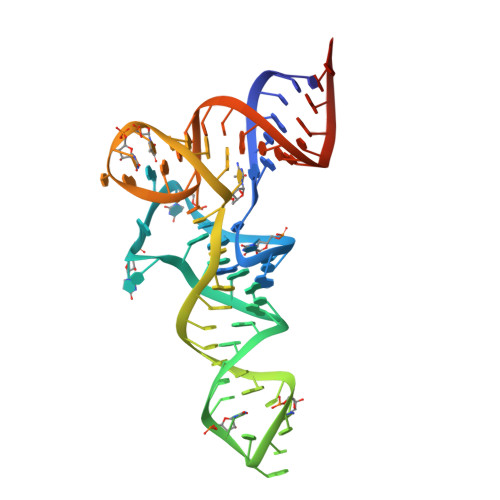
Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals.
Westhof, E., Dumas, P., Moras, D.(1988) Acta Crystallogr A 44: 112-123
- PubMed: 3272146
- Primary Citation of Related Structures:
2TRA, 3TRA, 4TRA - PubMed Abstract:
Four transfer RNA crystals, the monoclinic and orthorhombic forms of yeast tRNA(Phe) as well as forms A and B of yeast tRNA(Asp), have been submitted to the same restrained least-squares refinement program and refined to an R factor well below 20% for about 4500 reflections between 10 and 3 A. In yeast tRNA(Asp) crystals the molecules exist as dimers with base pairings of the anticodon (AC) triplets and labilization of the tertiary interaction between one invariant guanine of the dihydrouridine (D) loop and the invariant cytosine of the thymine (T) loop (G19-C56). In yeast tRNA(Phe) crystals, the molecules exist as monomers with only weak intermolecular packing contacts between symmetry-related molecules. Despite this, the tertiary folds of the L-shaped tRNA structures are identical when allowance is made for base sequence changes between tRNA(Phe) and tRNA(Asp). However, the relative mobilities of two regions are inverse in the two structures with the AC loop more mobile than the D loop in tRNA(Phe) and the D loop more mobile than the AC loop in tRNA(Asp). In addition, the T loop becomes mobile in tRNA(Asp). The present refinements were performed to exclude packing effects or refinement bias as possible sources of such differential dynamic behavior. It is concluded that the transfer of flexibility from the anticodon to the D- and T-loop region in tRNA(Asp) is not a crystal-line artefact. Further, analysis of the four structures supports a mechanism for the flexibility transfer through base stacking in the AC loop and concomitant variations in twist angles between base pairs of the anticodon helix which propagate up to the D- and T-loop region.
- Institut de Biologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique, Strasbourg, France.
Organizational Affiliation: