Differential mode of D-AKAP2 to PKA isoforms: Insights from D-AKAP2 PKA RII PDZK1 ternary complex structure
Sarma, G.N., Phan, R.H., Sankaran, B., Taylor, S.S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | 87 | Homo sapiens | Mutation(s): 0 Gene Names: PDZK1, CAP70, NHERF3, PDZD1 | 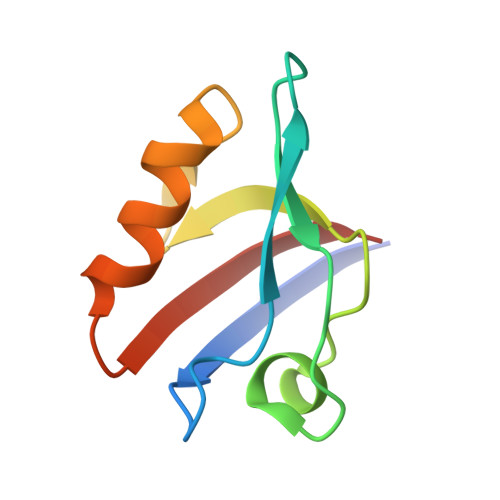 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q5T2W1 (Homo sapiens) Explore Q5T2W1 Go to UniProtKB: Q5T2W1 | |||||
PHAROS: Q5T2W1 GTEx: ENSG00000174827 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5T2W1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-dependent protein kinase type II-alpha regulatory subunit | 48 | Rattus norvegicus | Mutation(s): 0 Gene Names: Prkar2a | 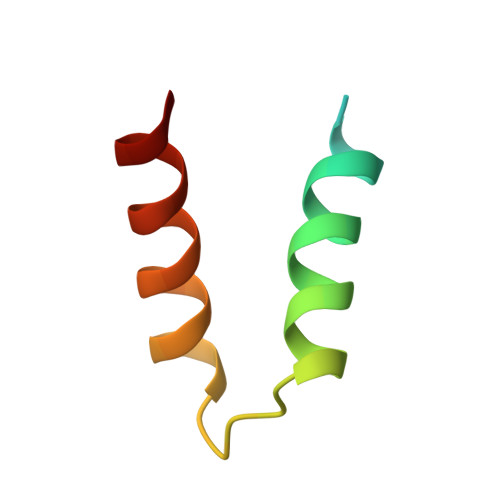 | |
UniProt | |||||
Find proteins for P12368 (Rattus norvegicus) Explore P12368 Go to UniProtKB: P12368 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12368 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| A-kinase anchor protein 10, mitochondrial | D, H, J [auth L] | 45 | Homo sapiens | Mutation(s): 0 Gene Names: AKAP10 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O43572 (Homo sapiens) Explore O43572 Go to UniProtKB: O43572 | |||||
PHAROS: O43572 GTEx: ENSG00000108599 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O43572 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 121.472 | α = 90 |
| b = 121.472 | β = 90 |
| c = 88.282 | γ = 120 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |